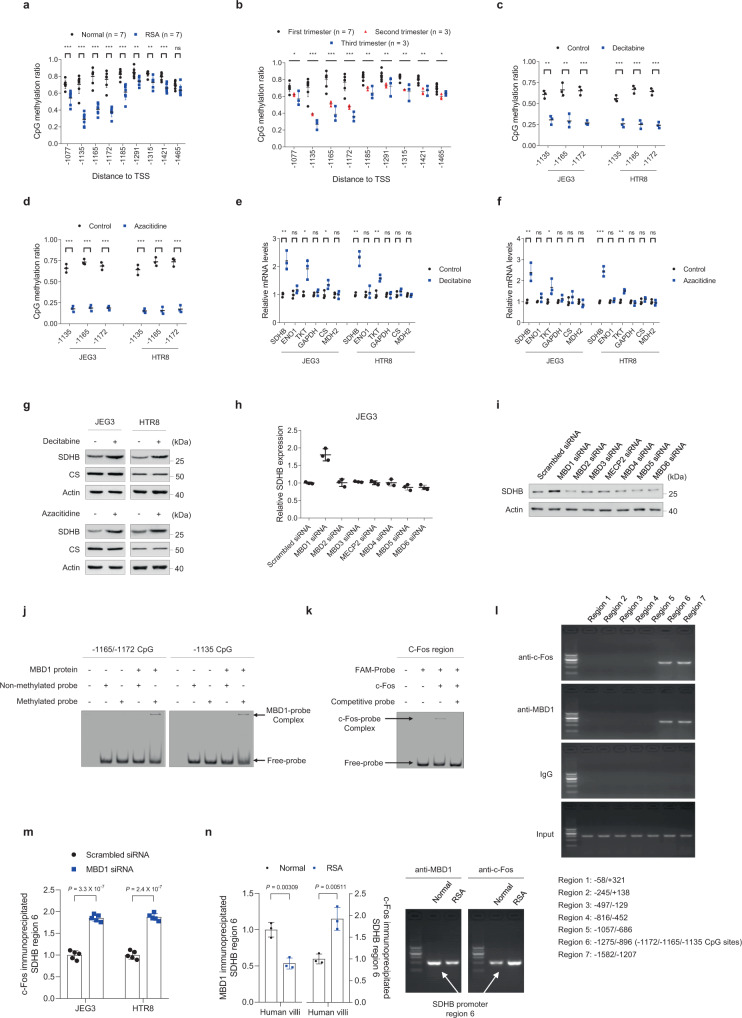
Fig. 3. DNA methylation regulates SDHB expression.
a CpG methylation levels in the SDHB promoter region of villi from individuals with RSA and normal controls (n = 7 persons/group). Exact P values in a are listed below: [−1077] 0.000624, [−1135] 2.5 × 10−6, [−1165] 7.6 × 10−7, [−1172] 9.9 × 10−7, [−1185] 0.000193, [−1291] 0.00272, [−1315] 0.00234, [−1421] 9.6 × 10−5, and [−1465] 0.0625. Other CpG sites besides those presented here are shown in Supplementary Fig. 2c. b SDHB promoter CpG methylation levels in villi from normal controls in different trimesters (n = 7 persons of the first trimester; n = 3 persons of the second trimester; n = 3 persons of the third trimester). Exact P values in b are listed below: [−1077] 0.00489, [−1135] 2.2 × 10−5, [−1165] 2.7 × 10−5, [−1172] 2.2 × 10−5, [−1185] 0.000316, [−1291] 0.00575, [−1315] 0.00011, [−1421] 0.000128 and [−1465] 0.00521. Other CpG sites besides those presented here are shown in Supplementary Fig. 2d. c, d CpG methylation levels in JEG3 and HTR8 cells treated with decitabine (c) (n = 3 independent experiments) or azacitidine (d) (n = 3 independent experiments). Exact P values in c are listed below: JEG3: [−1135] 0.00158, [−1165] 0.00463, and [−1172] 0.000308; HTR8: [−1135] 0.000877, [−1165] 0.000448, and [−1172] 0.000206. Exact P values in d are listed below: JEG3: [−1135] 0.000164, [−1165] 3.8 × 10−5, and [−1172] 7.4 × 10−5; HTR8: [−1135] 0.000202, [−1165] 1.0 × 10−5, and [−1172] 0.000112. e, f Gene expression in JEG3 and HTR8 cells treated with decitabine (e) (n = 3 independent experiments) or azacitidine (f) (n = 3 independent experiments). Exact P values in e are listed below: JEG3: SDHB 0.00377, ENO1 0.181, TKT 0.0112, GAPDH 0.339, CS 0.0436, and MDH2 0.973; HTR8: SDHB 0.00112, ENO1 0.559, TKT 0.00433, GAPDH 0.933, CS 0.283, and MDH2 0.763. Exact P values in f are listed below: JEG3: SDHB 0.000527, ENO1 0.171, TKT 0.0362, GAPDH 0.204, CS 0.303, and MDH2 0.398; HTR8: SDHB 0.000527, ENO1 0.383, TKT 0.00184, GAPDH 0.349, CS 0.271, and MDH2 0.638. g SDHB protein levels in cells treated with decitabine or azacitidine. h, i SDHB mRNA (h) (n = 3 independent experiments) and protein levels (i) in cells with knockdown of each methyl-CpG-binding protein. The knockdown efficiencies for each siRNA are shown in Supplementary Fig. 3b, c. j Binding abilities between MBD1 and double-strand DNA in the −1165/−1172 or −1135 CpG sites in EMSA. k Binding ability between c-Fos and double-strand DNA in the −1165/−1135 CpG sites in EMSA. l ChIP followed by PCR showing c-Fos and MBD1 occupancy at the SDHB promoter in cultured JEG3 cells. m ChIP-qPCR shows c-Fos occupancy at the SDHB promoter in MBD1-knockdown cells and control cells (n = 5 independent experiments). n ChIP followed by PCR/qPCR shows c-Fos and MBD1 occupancy at the SDHB promoter in villi from individuals with RSA (n = 3 biologically independent samples) and normal controls (n = 3 biologically independent samples). The left panel shows the ChIP-qPCR results. The right panel shows a representative result of ChIP-PCR. a–f, h, m, n Data represent the mean ± standard error, *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant. Two-tailed unpaired Student’s t tests in a, c–f, h, m, n; one-way ANOVA in b. Source data are provided as a Source Data file.