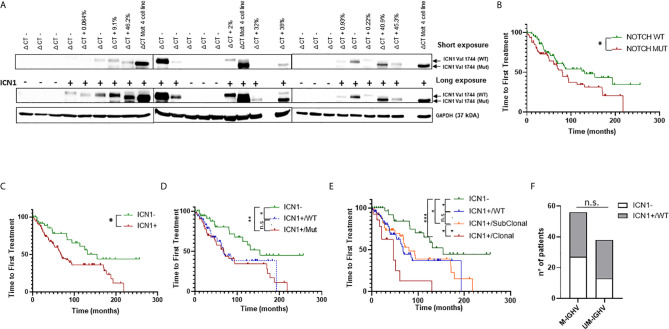
Figure 1.
Analysis of NOTCH1 activation and mutations in CLL patients. (A) Western blot analysis of NOTCH1 activation in whole cell lysates from 163 primary CLL cells. Short and long exposure of NOTCH1 activation (ICN1−/+) in 24 representative NOTCH1 WT (ΔCT−) and NOTCH1 mutated (ΔCT+, with relative AFs) CLL cases. ICN1 Val 1744 (WT) indicates the ICN1 NOTCH1 WT band, whereas ICN1 Val 1744 (Mut) indicates the ICN1 NOTCH1 mutated band (the difference in molecular weight is due to the presence of the deletion of a portion of PEST1). The Molt4 cell line was used as positive control. Protein loading was assessed by reprobing the blots with an anti-GAPDH antibody. (B) Kaplan–Meier analysis was used to determine TTFT in NOTCH1 mutated patients (NOTCH1 MUT) (n = 70) compared to NOTCH1 WT patients (n = 93) (80 vs 131 months, p = 0.04). Kaplan–Meier analysis was used to determine the influence on TTFT of the NOTCH1 activation and mutation status. Time to first treatment analysis according to: (C) NOTCH1 activation status (ICN1−, n = 43 and ICN1+, n = 120); (D) NOTCH1 activation status (ICN1−, n = 43) and NOTCH1 mutational status: ICN1+/WT (n = 57) and ICN1+/Mut (n = 63); (E) NOTCH1 activation status (ICN1−, n = 43) and NOTCH1 mutational status: ICN1+/WT (n = 57), ICN1+/Clonal mutated (n = 16) and ICN1+/Subclonal mutated (n = 47). (F) Stacked bar graph shows the IGHV mutational status in CLL patients with ICN1− compared to ICN1+/WT (Fisher’s exact Test). ***p < 0.001; **p < 0.01; *p < 0.05; and n.s., not significant.