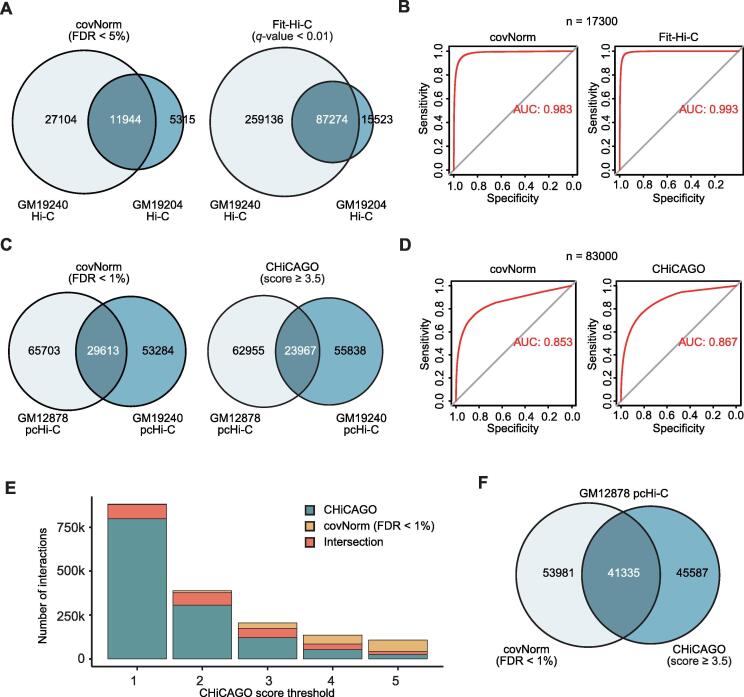
Fig. 4.
Evaluation of significant interaction calling from Hi-C and pcHi-C. A. Venn diagrams showing the fraction of reproducible significant interactions between GM19204 and GM19240 Hi-C data (Hypergeometric p-values ~0). B. ROC plots showing the accuracy of the covNorm and Fit-Hi-C for discovering GM19204′s significant interactions (n = 17300, nearest integer to the actual value) in the GM19240. C. Venn diagrams showing the fraction of reproducible significant interactions between GM12878 and GM19240 pcHi-C data (Hypergeometric p-values ~0). D. ROC plots showing the accuracy of the covNorm and CHiCAGO for discovering GM19240′s significant interactions (n = 83000, nearest integer to the actual value) in the GM12878. E. A stacked barplot showing the number of called interactions between covNorm (fixed threshold: FDR < 1%) and CHiCAGO (score ≥ 1 to 5, x-axis). Intersections between the two methods are marked with orange. F. A Venn diagram showing similar number of significance interaction call of covNorm (FDR < 1%) and CHiCAGO (score ≥ 3.5) in GM12878 pcHi-C data (Hypergeometric p-value ~0). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)