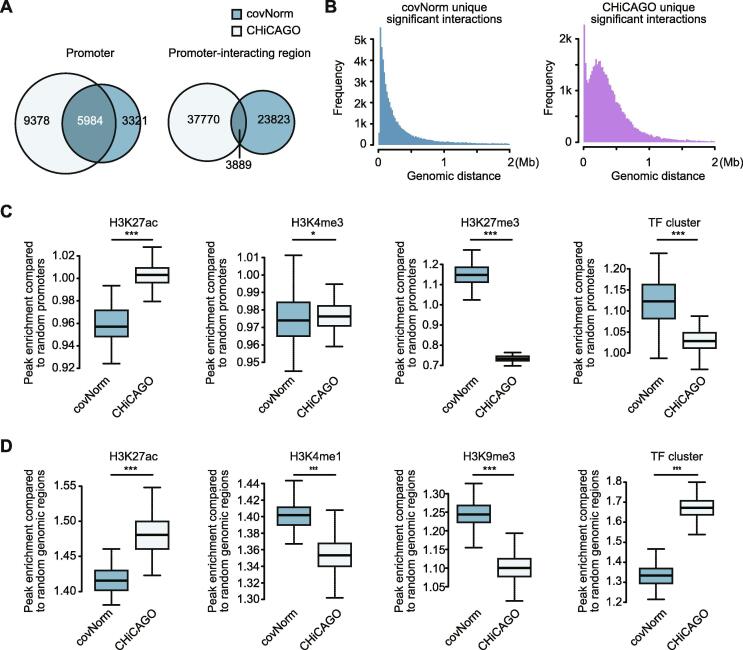
Fig. 5.
Different properties of method-specific significant interactions. A. Venn diagrams showing the fraction of overlaps of the unique promoter (left) and promoter-interacting region (right) between two methods’ specific significant interaction sets (Hypergeometric p-values > 0.05 for the promoter and < 9.56e-231 for the other regions). B. Histograms showing genomic distance distribution of method-specific significant interactions (Left: covNorm, right: CHiCAGO). C. Boxplots showing enriched chromatin signatures (H3K27ac, H3K4me3, H3K27me3, and TF cluster in series) at the promoter regions specifically associated with method-specific significant long-range chromatin contacts. The y-axis indicates the peak inclusion ratio compared to the promoter side fragment of random non-significant interactions. (Asterisks indicate significance, two-sided Komologv-Sminorv test, p-values of 4.807e-14, 0.01581, < 2.2e-16, and 1.071e-06 in series) D. Boxplots showing enriched chromatin signatures (H3K27ac, H3K4me1, H3K9me3, and TF cluster in series) of method-specific promoter-interacting region sets. The y-axis indicates the peak inclusion ratio compared to the promoter-interacting region side fragment of random non-significant interactions (Asterisks indicate significance, two-sided KS-test, p-value < 2.2e-16 for all boxplots). For the boxplots, the box represents the interquartile range (IQR) and the whiskers correspond to the highest and lowest points within 1.5 × IQR.