The genus Lyssavirus includes rabies virus as well as multiple diverse and recently described novel species. Using next-generation sequencing technologies, we have obtained the whole-genome sequence of Matlo bat lyssavirus, which was isolated from a Natal long-fingered bat (Miniopterus natalensis) in South Africa.
ABSTRACT
The genus Lyssavirus includes rabies virus as well as multiple diverse and recently described novel species. Using next-generation sequencing technologies, we have obtained the whole-genome sequence of Matlo bat lyssavirus, which was isolated from a Natal long-fingered bat (Miniopterus natalensis) in South Africa.
ANNOUNCEMENT
Lyssaviruses are bullet-shaped, with an approximately 12-kb-long negative-sense RNA genome encoding five proteins, namely, nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and an RNA-dependent polymerase (L). Recently, the lyssavirus species described have significantly increased, with 17 recognized lyssavirus species divided into two distinct phylogroups and several ungrouped viruses (1, 2).
Matlo bat lyssavirus (MBLV) was first detected in the brain of a Natal long-fingered bat (Miniopterus natalensis) on 2 September 2015 in Limpopo, South Africa, during targeted bat lyssavirus surveillance (3). Initial analyses of the PCR-amplified nucleoprotein gene sequence (1,353 nucleotides [nt]) indicated that MBLV is most closely related to West Caucasian bat virus (WCBV) (accession number NC_025377), with 80.9% nucleotide identity. Full-genome sequencing was performed using targeted amplicon sequencing on the MiSeq platform (Illumina). The virus was isolated using the standard mouse inoculation test (4). Viral RNA was extracted from mouse brains using TRIzol (Invitrogen); this was followed by first-strand cDNA synthesis using random hexamer primers (Integrated DNA Technologies) and the SuperScript III synthesis system (Thermo Fisher Scientific) (4, 5). The full genome was amplified using four different primer sets (Table 1) and the Phusion high-fidelity DNA polymerase system (New England Biolabs). Amplicons were purified using the Zymoclean gel DNA recovery kit (Zymo Research Corp.) followed by next-generation sequencing (NGS) for each individual purified amplicon using the Nextera XT library preparation kit (Illumina) on a MiSeq sequencing platform (Illumina), with paired-end read lengths of 2 × 300 bp and with 448× coverage obtained. All methods were according to the manufacturers’ instructions unless indicated otherwise. A total of 1,919,813 reads were obtained and assembled using the de novo assembly algorithm in CLC Main Workbench v12.0.2 (CLCbio). The entire genome was obtained apart from the 5′ and 3′ termini (71 nt and 50 nt, respectively), which were obtained by RNA circularization, cloning, and sequencing as described previously (5). The assembled viral reads and genomic terminal sequences were mapped to WCBV (GenBank accession number NC025377) as a reference sequence. The consensus sequence obtained was imported into the NCBI open reading frame (ORF) finder, where gene assignment was determined (6). All tools were run with default parameters unless otherwise specified. University of Pretoria Animal Ethics research approval was obtained (approval number EC054-14).
TABLE 1.
Primers designed for use during MiSeq amplicon sequencing of MBLV
| Primer set and name | Primer binding positiona | Sequence (5′ to 3′) |
|---|---|---|
| Set 1 | ||
| JW12 (7) | 55–74 | ATG TAA CAC CYC TAC AAT G |
| CG LYSSA S1 R | 4031–4049 | TRA ACA DBC CTC TYT CAT C |
| Set 2 | ||
| CG LYSSA S2 F | 2771–2793 | TCT GGB AAY MGA MGR ATG ATA GG |
| CG LYSSA S2 R | 6941–6967 | AYT TTT TCA TAT GGA CTT GAT CRT AMA |
| Set 3 | ||
| CG LYSSA S3 F | 6347–6364 | TRG AYT GGG ATG ARG ARA |
| CG LYSSA S3 R | 9569–9587 | CTV ACW GAG ATA TGA GAC A |
| Set 4 | ||
| CG LYSSA S4 F | 8671–8690 | GAG GAY CCW ACC ACH CTS AA |
| 001lys (8) | 1–15 | ACG CTT AAC GAM AAA |
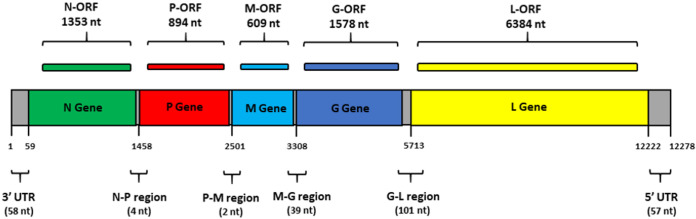
The genomic organization of MBLV was found to be similar to that of known lyssaviruses, with a complete genome length of 12,278 nt (GC content, 41.85%) (Fig. 1). MBLV was found to be most similar to WCBV, with a complete genome similarity of 78.9%. Gene positions and lengths were identified with the NCBI ORFfinder and subsequently compared to known lyssavirus sequences. Gene lengths and intergenic spacers (IGSs) were found to be similar to those of the unclassified lyssaviruses, with the G-L IGS (101 nt) being significantly larger than those of phylogroup I and II lyssaviruses. The level of similarity of known antigenic regions between MBLV and the other unclassified lyssaviruses also provided initial evidence that cross neutralization will most probably not occur when immunized hosts are exposed to this virus.
FIG 1.
Genome characteristics determined for MBLV. The N, P, M, G, and L protein gene lengths are shown. IGS and 5′ and 3′ untranslated region (UTR) lengths are indicated in parentheses.
Data availability.
The complete genome sequence of MBLV has been deposited in GenBank under accession number MW653808. Raw NGS sequence reads have been deposited in the SRA under accession number PRJNA708137.
ACKNOWLEDGMENTS
Funding was from the South African Research Chair Initiative (held by W.M.) of the Department of Science and Innovation and was administered by the National Research Foundation of South Africa (grant 98339). The National Research Foundation funded the equipment based at the DNA Sanger sequencing facility in the Faculty of Natural and Agricultural Sciences, University of Pretoria (grant 78566). Student funding was received from the National Research Foundation (grant 98339) and the Poliomyelitis Research Foundation (grant 18/54).
REFERENCES
- 1.Markotter W, Coertse J. 2018. Bat lyssaviruses. Rev Sci Tech 37:385–400. doi: 10.20506/rst.37.2.2809. [DOI] [PubMed] [Google Scholar]
- 2.International Committee on Taxonomy of Viruses. 2020. Virus taxonomy: 2019 release. https://talk.ictvonline.org/taxonomy.
- 3.Coertse J, Grobler CS, Sabeta CT, Seamark EC, Kearney T, Paweska JT, Markotter W. 2020. Lyssaviruses in insectivorous bats, South Africa, 2003–2018. Emerg Infect Dis 26:3056–3060. doi: 10.3201/eid2612.203592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupprecht CE, Fooks AR, Abela-Ridder B. 2018. Laboratory techniques in rabies, 5th ed, vol 1, p 74–83. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Kuzmin IV, Wu X, Tordo N, Rupprecht CE. 2008. Complete genomes of Aravan, Khujand, Irkut and West Caucasian bat viruses, with special attention to the polymerase gene and non-coding regions. Virus Res 136:81–90. doi: 10.1016/j.virusres.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 6.NCBI. 2021. Open reading frame finder. https://www.ncbi.nlm.nih.gov/orffinder.
- 7.Heaton PR, JohnstonE P, McElhinney LM, Cowley R, O'Sullivan E, Whitby JE. 1997. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. J Clin Microbiol 35:2762–2766. doi: 10.1128/JCM.35.11.2762-2766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markotter W, Kuzmin I, Rupprecht CE, Randles J, Sabeta CT, Wandeler AI, Nel LH. 2006. Isolation of Lagos bat virus from water mongoose. Emerg Infect Dis 12:1913–1918. doi: 10.3201/eid1212.060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of MBLV has been deposited in GenBank under accession number MW653808. Raw NGS sequence reads have been deposited in the SRA under accession number PRJNA708137.