Figure 1.
LELNs enhance lactobacilli survival in the small intestine by enhancing bile resistance
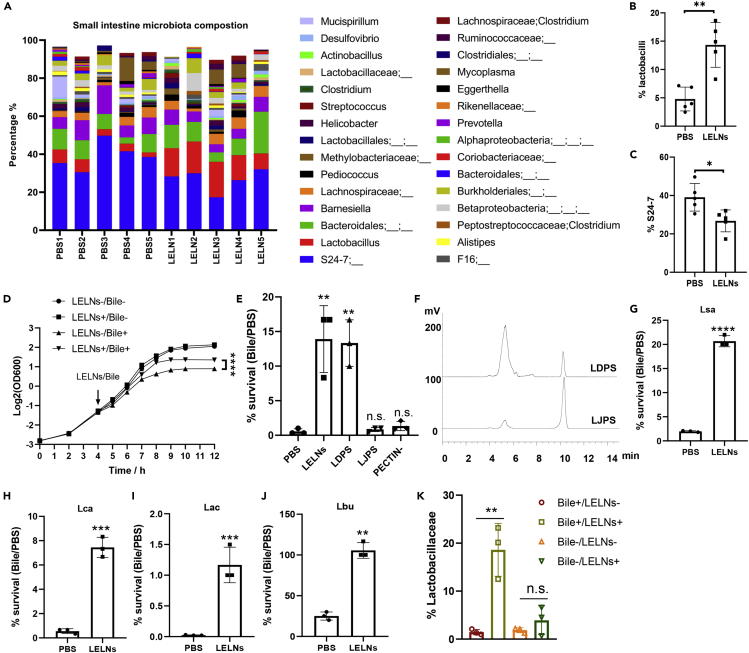
(A) 16S-rDNA sequencing analysis of small intestine microbiota composition in LELN-treated and PBS control C57BL/6J mice (n = 5). Top 30 genera in the small intestine were shown.
(B and C) Statistical data of Lactobacillus and S24-7 percentage in the small intestine microbiota.
(D) LGG growth curve under different treatments. LELN- and LELN+ indicate LGG growth without or with LELN treatment, and Bile- and Bile+ indicate LGG growth without or with bile treatment.
(E) LGG survival rate in a bile challenge, 1 × 1010/mL LELNs and PECTIN- or 10 mg/mL LDPS (comparable with 5 × 1010/mL LELNs) and LJPS were used in the experiments.
(F) Comparative analysis of polysaccharide composition of LDPS and LJPS by HPLC; 0.1 mg polysaccharides was injected into the HPLC system.
(G–J) Bile resistance test of four different lactobacilli strains as indicated with or without LELN treatment. Lsa represents Lactobacillus salivarius LS-33, Lca represents Lactobacillus casei LC-11, Lac represents Lactobacillus acidophilus 4356, Lbu represents Lactobacillus bulgaricus LB-87.
(K) Percentage of Lactobacillaceae in the small intestine microbiota after 24 h in vitro culture with or without LELNs treatment. The data are presented as values with standard deviation (mean ± SD). The significance is shown as ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; p > 0.05 was considered not significant (n.s.).