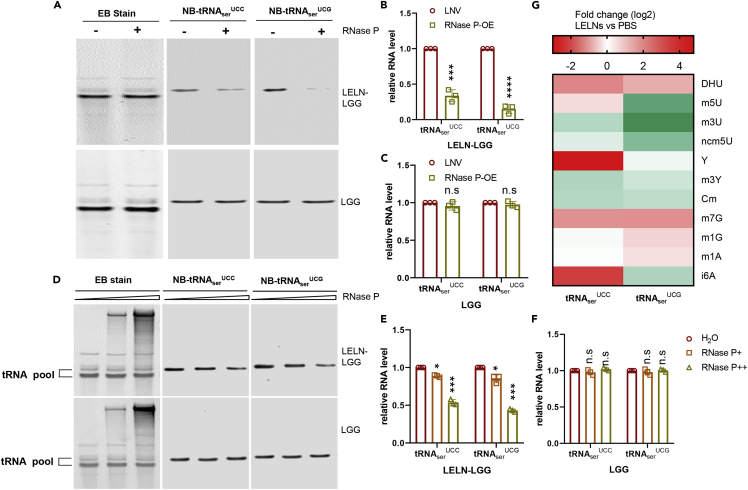
Figure 4.
RNase P is responsible for LELNs-induced tRNAserUCC and tRNAserUCG decay in LGG
(A–C) tRNAserUCC and tRNAserUCG levels in LGG after RNase P overexpression. tRNAserUCC and tRNAserUCG levels from LGG (bottom panels) or LELN-LGG (top panels) were analyzed by northern blot (A) and quantified by ImageJ software (B and C). ‘+’ indicates RNase P delivered by LNV, ‘-’ indicates LNV control without RNA included. EB stain was used as the loading control.
(D–F) In vitro RNase P cleavage assay of tRNAserUCC and tRNAserUCG derived from LGG or LELN-LGG. Two different doses of RNase P, 200 ng (middle lanes or RNase P+), and 2 μg (right lanes or RNase P++) were used in the cleavage assay. H2O was used as a control (left lanes); EB stain was used as a loading control.
(G) Nucleoside modifications analysis of tRNAserUCC and tRNAserUCG by HPLC-MASS, the nucleoside modification levels are shown as fold changes due to LELN treatment. The data are presented as values with standard deviation (mean ± SD). The significance is shown as ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 and p > 0.05 was considered not significant (n.s.).