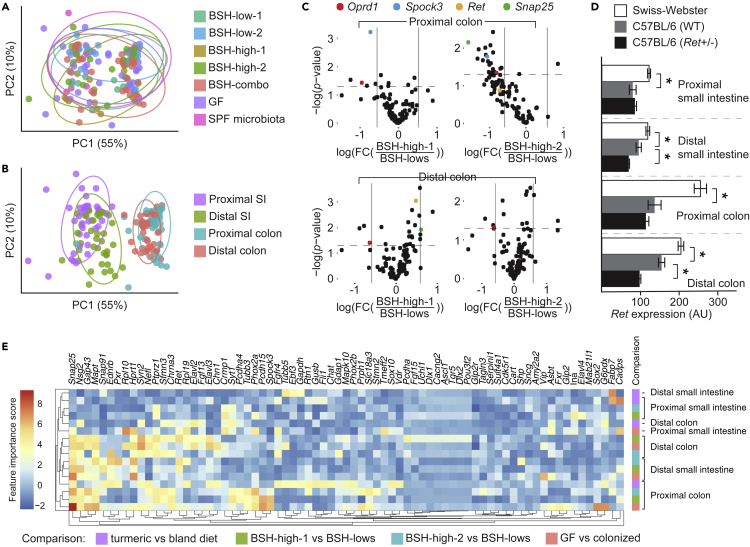
Figure 6.
Gene expression signatures were microbiota-specific, highly reproducible, and distinct between gut segments (See also Figures S5 and S7 and Table 1)
In an analysis of Bray-Curtis dissimilarities between ENS-specific gene expression signatures of intestinal samples in our dataset, we observed significant clustering by (A) microbiota and (B) biogeography.
(C) Volcano plots for BSH-high vs BSH-low in the colon. The y axis reflects log10-transformed p values, and the x axis shows log2-transformed fold-changes.
(D) Ret expression as a function of genotype.
(E) The most discriminatory features in comparisons of 4 dietary and microbial exposures as determined using a Random Forest classifier, filtered for genes found to have an importance score of >1 in at least 1 comparison, with hierarchical clustering applied across rows (specific comparison) and columns (genes).
In (D), mean values ± SEM are shown.