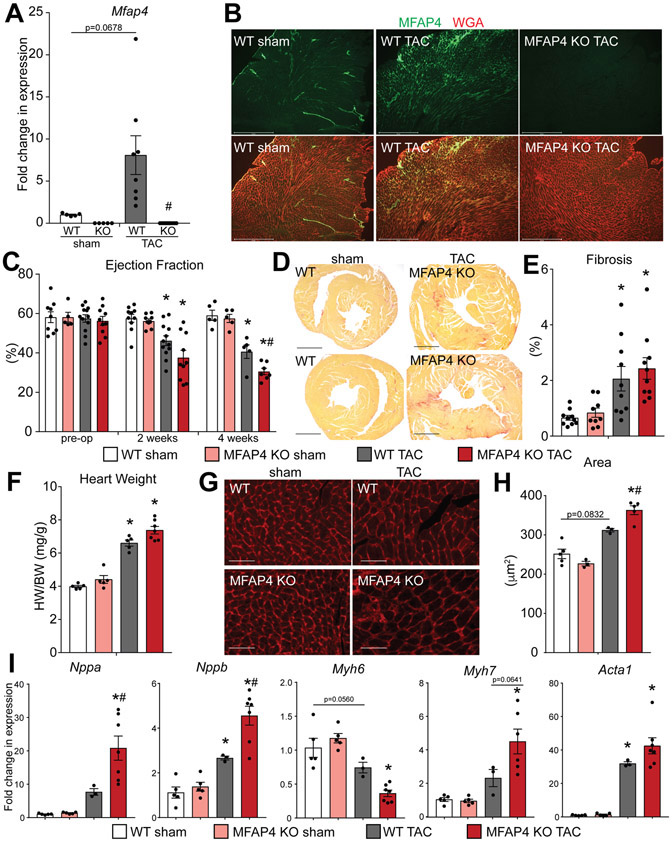
Figure 2. MFAP4 KO mice demonstrate cardiac dysfunction and altered cardiac remodeling following chronic pressure overload.
A. qPCR analysis for Mfap4 relative to Rpl7 in the indicated groups. (WT and KO sham n=5, WT TAC n=8, KO TAC n=12). (Two-way ANOVA results: WT sham vs TAC p=0.0678; WT vs KO TAC #p=0.00075). B. Immunofluorescence staining of MFAP4 (green) and wheat germ agglutinin (WGA; red) in hearts from the indicated groups. Scale bar, 500 μm. C. Echocardiographic quantification of percentage ejection fraction in WT and MFAP4 KO mice prior to surgery (pre-op) and every two weeks after induction of pressure overload. (WT sham pre-op n=9, KO sham pre-op n=5, WT TAC pre-op n=12, KO TAC pre-op n=10; WT sham 2 weeks n=10, KO sham 2 weeks n=9, WT TAC 2 weeks n=12, KO TAC 2 weeks n=10; WT sham 4 weeks n=5, KO sham 4 weeks n=5, WT TAC 4 weeks n=5, KO TAC 4 weeks n=7). (Two-way ANOVA results for 2 wks time point: WT sham vs TAC *p=0.0112; KO sham vs TAC *p=0.000175; 4 wks time point: WT sham vs TAC *p=0.00042; KO sham vs TAC *p=0.0000012; WT vs KO TAC #p=0.0421). D. Representative picrosirius red – stained cardiac cross-sections from WT and MFAP4 KO mice following 4 weeks of sham or pressure overload (TAC) surgery. Scale bar, 1 mm. E. Quantification of percent fibrosis from picrosirius red - stained cardiac cross-sections in the indicated groups after sham or pressure overload (TAC) surgery. (WT and KO sham n=5, WT TAC n=5, KO TAC n=7). (Two-way ANOVA results: WT sham vs TAC *p=0.0079; KO sham vs TAC *p=0.0060). F. Gravimetric analysis of heart weight to body weight ratio (HW/BW) in the indicated groups 4 weeks after sham or pressure overload (TAC) surgery. (Two-way ANOVA results: WT sham vs TAC *p=0.000017; KO sham vs TAC *p=0.0000086). G. Representative images of WGA-stained cardiac cross-sections of WT or MFAP4 KO mice 4 weeks after sham or pressure overload (TAC) surgery. Scale bar, 50 μm. H. Quantification of cardiomyocyte cross-sectional area based on WGA-stained cardiac cross-sections in the indicated groups. (WT sham n=5, KO sham n=3, WT TAC n=3, KO TAC n=5). (Two-way ANOVA results: WT sham vs TAC p=0.0832; KO sham vs TAC *p=0.00019; WT vs KO TAC #p=0.0420). I. qPCR analysis for hypertrophic gene expression relative to Rpl7 in the indicated groups. (WT and KO sham n=5, WT TAC n=3, KO TAC n=7). (Two-way ANOVA results - Nppa: KO sham vs TAC *p=0.000030; WT vs KO TAC #p=0.0037. - Nppb: WT sham vs TAC *p=0.0201; KO sham vs TAC *p=0.0000098; WT vs KO TAC #p=0.0022. - Myh6: WT sham vs TAC p=0.0560; KO sham vs TAC *p=0.00011. - Myh7: KO sham vs TAC *p=0.000087; WT vs KO TAC p=0.0641. - Acta1: WT sham vs TAC *p=0.0037; KO sham vs TAC *p=0.000055).