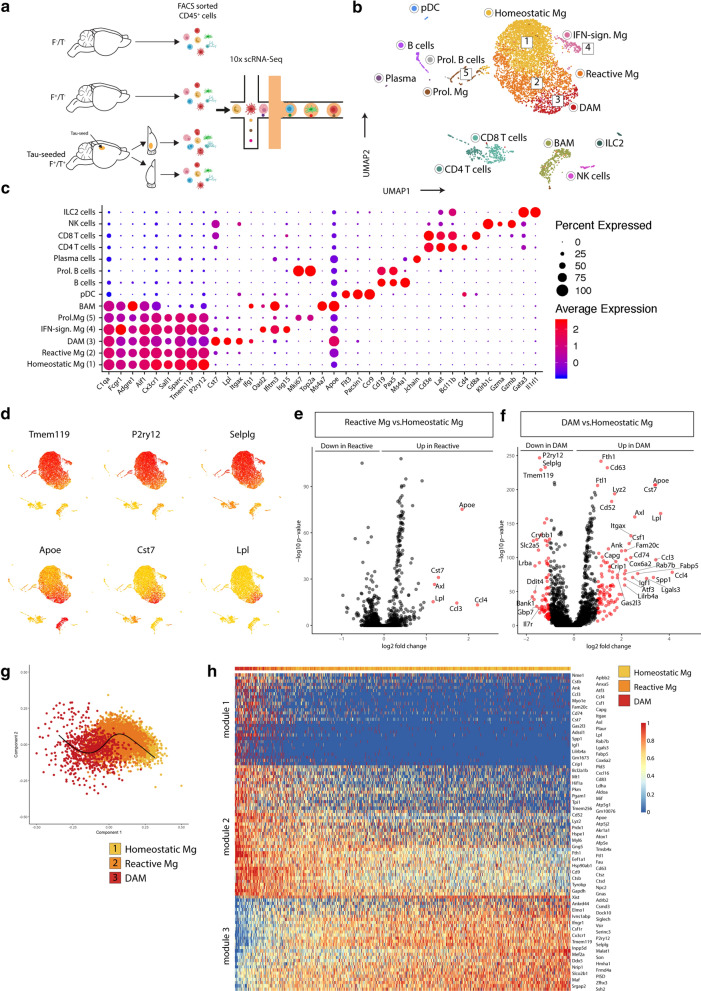
Fig. 4.
scRNA-Seq analysis of F−/T−, F+/T− and tau-seeded F+/T+ brains reveals microglial activation and progression towards a DAM state. a Schematic overview of the 10 × chromium scRNA-Seq setup used on brains from F−/T−, F+/T−, and tau-seeded F+/T+ mouse models. Brains were collected from n = 4 mice for each condition. b UMAP-projection containing 842 F−/T− cells, 2666 F+/T− cells, and 1500 ipsilateral and 1500 contralateral tau-seeded F+/T+ cells which were randomly downsampled for visualization purposes in the UMAP plot. Clusters 1 to 5 correspond to different activation states of microglia populations. 1: homeostatic microglia, 2: reactive microglia, 3: DAM, 4: IFN-sign. microglia, 5: proliferative microglia. c Dot plot visualizing expression of key marker genes for each of the clusters that were identified in (b). d UMAP plots showing the expression of homeostatic microglia marker genes (top) and DAM marker genes (bottom). e Volcano plot showing genes that are DE between reactive microglia and homeostatic microglia. f Volcano plot showing genes that are DE between homeostatic microglia and DAM. DE threshold: -log10(adjusted p) > 5, log2(FC) > 1; p value adjustment was performed using Bonferroni correction (e–f). g SCORPIUS trajectory inference was run on microglia clusters 1, 2 and 3 from (b). Cells were automatically ordered along a linear trajectory and colored according to the cluster colors visualized in (b). h The top 100 genes that were found to define the trajectory in panel g, were clustered into three gene modules (normalized expression) that are down-or upregulated as homeostatic microglia transition towards DAM