ABSTRACT
The present study is an attempt to determine the lipid composition of Candida auris and to highlight if the changes in lipids can be correlated to high drug resistance encountered in C. auris. For this, the comparative lipidomics landscape between drug-susceptible (CBS10913T) and a resistant hospital isolate (NCCPF_470033) of C. auris was determined by employing high throughput mass spectrometry. All major groups of phosphoglycerides (PGL), sphingolipids, sterols, diacylglycerols (DAG) and triacylglycerols (TAG), were quantitated along with their molecular lipid species. Our analyses highlighted several key changes where the NCCPF_470033 showed an increase in PGL content, specifically phosphatidylcholine, phosphatidylglycerol, phosphatidylserine, phosphatidylinositol, and phosphatidylethanolamine; odd chain containing lipids and accumulation of 16:1-DAG and 16:0-DAG; depletion of 18:1-TAG and 18:0-TAG. The landscape of molecular species displayed a distinct imprint between isolates. For example, the levels of unsaturated PGLs, contributed by both odd and even-chain fatty acyls were higher in resistant NCCPF_470033 isolate, resulting in a higher unsaturation index. Notwithstanding, several commonalities of lipid compositional changes between resistant C. auris and other Candida spp., the study could also identify distinguishable changes in specific lipid species in C. auris. Together, the data highlights the modulation of membrane lipid homeostasis associated with drug-resistant phenotype of C. auris.
Keywords: Lipids, pathogenic fungi, functions, mass spectrometry
The present study determines the lipid composition of Candida auris and highlights alterations in lipids that may be correlated to high drug resistance in fungi.
INTRODUCTION
The scope of diseases caused by human fungal pathogens ranged from superficial to chronic by a variety of Candida species. The prevalent human fungal infections are majorly caused by Candida albicans and non-albicans Candida (NAC) species. Apart from common fungal species, newer species of Candida are emerging as belligerent pathogens that display a high level of drug resistance (Brown et al. 2012; Fairlamb et al. 2017). In 2009 from Japan, a close relative of Candida haemulonii was identified and named as Candida auris, represents one such example (Satoh et al. 2009; Kathuria et al. 2015). Fungemia, due to C. auris, now prevalent worldwide, is associated with a high mortality rate and treatment failure, in addition to being highly resistant to commonly used antimycotics such as azoles, polyenes, and echinocandins (Chowdhary, Sharma and Meis 2017; Lockhart et al. 2017; Lone and Ahmad 2019).
The most common contributors involved in reducing the impact of antifungals used by other pathogenic fungi include overexpression of multidrug transporters (Kumari et al. 2018; Wasi et al. 2019) and drug target alterations also represent another common strategy that excludes the antifungals from reaching out to its target (Lupetti et al. 2002; Shor and Perlin 2015). Azoles target 14α-demethylase encoded by ERG11, while echinocandins target 1,3-β-glucan synthase encoded by FKS genes. Both the targets are widely reported to acquire mutations in drug-resistant isolates in Candida isolates (Lupetti et al. 2002; Hou et al. 2019). Notwithstanding, the well understood and widely documented features of drug resistance, microorganisms are continually developing new and varied ways of rendering generally used antifungal drugs. (Anderson 2005). Several recent reports have shown new resistance mechanisms that fungal cells resort to thwart drug effectiveness includes stress responses that are crucial in assuring the survival of the cell (Shor and Perlin 2015).
In Candida, the enzymes of lipid biosynthetic pathways represent a target of many known antifungal drugs (Prasad and Singh 2013). Previous independent studies on Candida by our group and others demonstrated that membrane lipids composition could influence the drug susceptibilities in Candida cells (Lattif et al. 2011; Singh, Yadav and Prasad 2012). Besides, the virulence of many pathogens is also impacted by lipids, the phospholipid synthesis genes, PSD1 and PDS2 play a crucial role in azole resistance, cell wall integrity and virulence of C. albicans (Gonçalves et al. 2017; Khandelwal et al. 2018). Moreover, any changes in the lipid homeostasis and membrane lipid asymmetric distributions between two monolayers of the plasma membrane also shown to affect drug resistance and virulence (Gonçalves et al. 2017; Khandelwal et al. 2018). Together, the drug susceptibilities depend on their diffusion rates, efflux by membrane pump proteins, and the local membrane environment (Balzi and Goffeau 1995; Prasad and Singh 2013).
Recent advances in high throughput mass spectrometry-based strategies for a complete analysis of cellular lipids have been developed and employed in fungi (Singh and Del Poeta 2016). By employing high throughput mass spectrometry approach, we have earlier characterized the lipidome of various Candida spp. to show that while all Candida species comprise similar levels of various phospholipid classes, they exhibit distinct molecular lipid species imprint (Singh et al. 2010). The comparative lipidomics study that followed between susceptible and resistant clinical genetically matched isolates of C. albicans, presented evidence that changes in lipid homeostasis can not only influence drug resistance phenotypes but also impacts the cell wall integrity and mitochondrial function (Singh and Prasad 2011; Singh, Yadav and Prasad 2012).
That the prevalence of highly drug-resistant clinical isolates of C. auris, which cannot be explained based on the contributions of established resistance mechanisms (Kim et al. 2019; Rybak et al. 2019), makes it imperative to seek new alternatives which could also be important in presenting such level of drug resistance. For this, the present study, using the ESI-MS/MS (electrospray ionization tandem mass spectrometry) approach, has focused on analyzing comparative lipidome between drug-resistant and susceptible C. auris isolates. Our analyses quantified five major lipid groups of C. auris, namely PGLs, sphingolipids (SL), sterols, diacylglycerols (DAG) and triacylglycerols (TAG) along with the composition of their molecular lipid species. The comparative lipid profiles of C. auris strain CBS10913T (drug-susceptible) and NCCPF_470033 (drug-resistant) revealed marked changes in their compositions, suggesting a major remodeling of polar lipids in drug resistant C. auris.
MATERIALS AND METHODS
Materials
Lipid standards were purchased from Sigma-Aldrich, USA and Avanti polar lipids Inc., USA. All solvents and chemicals were mass spectrometry grade and purchased from Sigma-Aldrich, USA and Fisher Scientific, UK.
Strains and culture conditions
Candida auris strain CBS10913T was purchased from CBS database, Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. The drug-resistant C. auris NCCPF_470033 strain (this study) is a hospital isolate from India and was obtained from the National Culture Collection of Pathogenic Fungi, Postgraduate Institute of Medical Education & Research, Chandigarh, India. Cultures were maintained on YEPD agar plates at 30°C. For lipid extraction, 106 cells (1.0 O.D.) were inoculated in 50 mL YEPD medium and grown for 6 hrs, and approximately 5 × 108 cells were harvested, washed with dH2O thrice before lipid extraction (Singh, Yadav and Prasad 2012).
Drug susceptibility assessment
Minimum inhibitory concentrations (MIC) of fluconazole (FLC), itraconazole (ITZ), ketoconazole (KTZ), miconazole (MCZ), amphotericin B (AmB), nystatin (NYS), caspofungin (CSP) and micafungin (MICA) against the C. auris strains were determined as described in method M27-A3 by the Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS, USA) (Pfaller et al. 2008).
Lipid extraction
Harvested cells were broken in methanol using glass beads (Glaperlon, 0.4–0.6 mm), and lipids were extracted by the Folch extraction method using chloroform: methanol (2:1, v/v), as described earlier (Folch, Lees and Sloane Stanley 1957). Lipid extracts were dried in N2 and kept in −20°C until analyzed. Lipid dry weights were recorded at this step for the normalization of mass spectral data.
Mass spectrometry analysis
Lipid extracts were dissolved in 1 mL chloroform. To the 2–3 µL aliquots of these dissolved lipids, internal standards were added as described previously (Singh et al. 2010; Singh and Prasad 2011; Singh, Yadav and Prasad 2012), and volume made up to 1.4 mL using chloroform: methanol: 300 mM ammonium acetate (100:5:26, v/v/v). Lipid extracts were then analyzed on Xevo TQ-XS triple quadrupole mass spectrometer (XEVO-TQS#WAA627, Waters, UK, Milford, MA). Levels of various lipid classes and individual molecular lipid species were determined using the neutral loss, negative and positive multiple precursor ion scans as described earlier (Singh et al. 2010; Singh and Prasad 2011; Singh, Yadav and Prasad 2012).Instrument parameters settings are listed in supplementary sheet S1. Data processing was performed using the TargetLynx XS™ software (Waters, UK, Milford, MA) and data normalized to lipid dry weight and represented as nmol/mg lipid dry weight. Lipid structures and their nomenclature were annotated according to previously published studies (Bowden et al. 2017) (www.lipidmaps.org, www.k-state.edu/lipid).
Gas chromatography mass spectrometry (GC-MS)
For free sterol analysis, base hydrolyzed lipid extract were derivatized with N,O-Bis (trimethylsilyl) trifluoroacetamide with trimethyl-chlorosilane (BSTFA/TMCS, Sigma, USA) and analyzed on DB5-MS column (30 m × 0.2 mm × 0.20 µm) as described previously (Singh, Yadav and Prasad 2012). The retention time and mass spectral patterns of external standards were used for identification of sterol species. For fatty acid (FA) analyses, base hydrolyzed lipid extract were derivatized with BF3-Methanol and analyzed on Omegawax column (30 m × 250 µm × 0.25 µm) as described previously (Singh et al. 2017). The retention time and mass spectral patterns of fatty acid methyl ester standard mix (Sigma, USA) were used as a reference to identify respective FAs (Sharma et al. 2012).
Statistical analysis
Experiments were performed in triplicates, and all the lipid data sets are represented as mean ± standard error mean (SEM). Statistical significance was determined using student's t-test and P-values < 0.05 were considered significant. The principal component analysis (PCA) was performed using the XLSTAT software (Addinsoft, US) on Microsoft Excel platform. PCA between C. auris strains CBS10913T and NCCPF_470033 was performed using the whole lipidome data sets (582 lipid species as nmol/mg lipid dry weight, supplementary sheet S2) as well as the phospholipid species data (144 lipid species as % of total PGL + SL). However, for comparative PCA of C. auris with other Candida strains, we have used the data for only phospholipid species (as % of total PGL + SL, supplementary sheet S3), which are reported in the previously published lipidomic studies by our group (Singh and Prasad 2011; Singh, Mahto and Prasad 2013). We chose the mole% data of 144 phospholipid species for comparative purposes as these were detectable in the current study as well as in the earlier published lipidomes of Candida.
RESULTS AND DISCUSSION
NCCPF_470033 strain is highly resistant to azoles
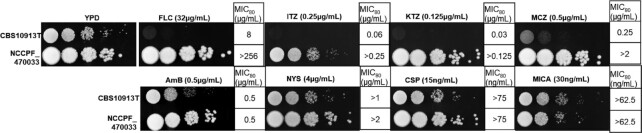
For the lipidome analysis, we selected two strains of C. auris; CBS10913T, which was highly susceptible to antifungals and resistant hospital isolate NCCPF_470033. The CBS10913T strain was susceptible to all the tested drugs that included azoles, polyenes, and echinocandins. However, in comparison with the susceptible CBS10913T strain, NCCPF_470033 isolate showed high resistance towards FLC (MIC80 > 256.0 µg/mL), ITZ (MIC80 > 0.25 µg/mL), KTZ (MIC80 > 0.125 µg/mL) and MCZ (MIC80 > 2.0 µg/mL) and showed no change in susceptibility towards AmB (MIC80 0.5 µg/mL), NYS (MIC80 > 2.0 µg/mL), CSP (MIC80 75 ng/mL) and MICA (MIC80 62.5 ng/mL) (Fig. 1). The differences in drug susceptibilities between CBS10913T and NCCPF_470033 isolates allowed us to make comparisons of their lipid profiles.
Figure 1.
Drug susceptibility profiling of CBS10913T and NCCPF_470033 strains. The panels show the drug susceptibility of C. auris strains on FLC, ITZ, KTZ, MCZ, AmB, NYS, CSP and MICA using the spot assays. The MIC values, as determined by the broth dilution method (Pfaller et al. 2008), are also depicted.
Azole resistant NCCPF_470033 show elevated levels of polar lipids
A comparative lipidomics was performed to underscore the importance of lipids in influencing drug resistance in C. auris. For this, lipids were extracted as described earlier from FLC susceptible CBS10913T and FLC resistant NCCPF_470033 strains. Lipid dry weights of extracts in triplicates obtained from 5 × 108 CBS10913T and NCCPF_470033 cells were 1.86 ± 0.03 and 2.23 ± 0.08 mg, respectively. The extracted lipids were subjected to mass spectrometry analysis on ESI-MS/MS (Electrospray Ionization mass spectrometry) and GC-MS as described earlier. In our analysis, 582 lipid species were detected belonging to five different lipid groups namely, PGLs, SLs, sterols, DAGs and TAGs. Moreover PGLs further divided into eight different lipid subclasses, namely lysophosphatidylcholine (LPC, 1-acyl-sn-glycero-3-phosphocholine) phosphatidylcholine (PC, 1,2-diacyl-sn-glycero-3-phosphocholine), lysophosphatidylethanolamine (LPE, 1acyl-sn-glycero-3-phosphoethanolamine), phosphatidylethanolamine (PE,1,2-diacyl-sn-glycero-3-phosphoethanolamine) phosphatidylinositol (PI, 1,2-diacyl-sn-glycero-3-phospho-(1'-myo-inositol), phosphatidylserine (PS, 1,2-diacyl-sn-glycero-3-phosphoserine), phosphatidylglycerol (PG, 1,2-diacyl-sn-glycero-3-phospho-(1'-sn-glycerol), phosphatidic acid (PA, 1,2-diacyl-sn-glycero-3-phosphate).
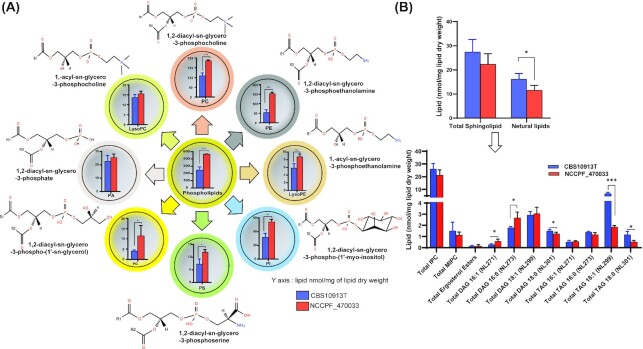
The quantitation of lipids revealed that the azole-resistant isolate NCCPF_470033 had 1.9 folds higher PGLs amounting to 467.6 ± 2.1 nmol/mg lipid dry weight when compared with the CBS10913T strain (Fig. 2A). This data coincided well with the increased levels of various PGL subclasses like PC, PE, LPE, PI, PS, and PG were as much as 2.9-fold higher in NCCPF_470033, compared to the CBS10913T strain. The contents of PA and LPC did not show any change between these isolates (Fig. 2A). PC was the most abundant PGL in the C. auris followed by PE. Interestingly PC was 1.7-fold higher in azole-resistant isolates, but PE showed the maximum change (2.9-fold) between the tested isolates among PGLs.
Figure 2.
The total content of different lipid classes between CBS10913T and NCCPF_470033 strains. (A), The total PGL content, along with the total class contents of PC, LPC PE, LPE, PI, PS, PG and PA, in CBS10913T and NCCPF_470033 strains of C. auris is depicted. (B), The total SL and neutral lipid contents, along with the total class contents of IPC, MIPC, DAG and TAG, in CBS10913T and NCCPF_470033 strains of C. auris is depicted. The DAG and TAG content were determined based on the attached FA chain, as described earlier, For example, NL271, NL273, NL299 and NL301 scans were used to detect 16:1, 16:0, 18:1 and 18:0 -FA containing DAG or TAG species, respectively. Data represents nmol per mg lipid dry weight and can be found in supplementary sheet S2. Mean ± SEM of 3 replicates are plotted. The P-values of < 0.05, <0.01 and < 0.001 are represented by *, ** and ***, respectively.
Our analysis of SLs was limited, and only two classes of SL were detected, which displayed no significant difference between the CBS10913T and NCCPF_470033 strains (Fig. 2B). The neutral lipids are the storage lipids and contain two major subclasses DAG and TAG. The total neutral lipid content was ∼39% lower in NCCPF_470033 compared to the CBS10913T strain (Fig. 2B). While we observed as much as 2.1 folds increase in 16:1-DAG and 16:0-DAG content of NCCPF_470033, there were as much as 3.6 folds decrease in 18:1-TAG and 18:0-TAG content of NCCPF_470033, compared to the CBS10913T strain (Fig. 2B). In corroboration to these data, our GC-MS analysis showed that the content of 16C FA was 8% higher, and the content of 18C FA was 45% lower in NCCPF_470033 compared to the CBS10913T strain (Supplementary Fig. S1). However, composition of ergosterol esters showed no significant difference between the two strains (Fig. 2B). A recent multiomics study of Zamith-Miranda et al. also reported that PC and TAG were most diverse lipid classes between resistant and sensitive C. auris isolates. However, lipids with unsaturation C16:1 FA was lower in resistant isolate (Zamith-Miranda et al. 2019). This is in accordance to our present data set of lipid profiles of resistant and susceptible C. auris isolates.
Molecular lipid species of PGLs show distinct imprint between susceptible and resistant strains
Alterations in membrane fluidity by changes in lipid homeostasis are known to affect drug resistance in Candida species (Mukhopadhyay, Kohli and Prasad 2002; Prasad et al. 2005). These alterations include changes in FA chain lengths, number of double bonds, hydroxylations, methylations, and backbone structure. We monitored these changes by quantifying the variations in molecular lipid species compositions between the CBS10913T and NCCPF_470033 strains. We identified and quantitated a total of 582 molecular species of various classes of lipids in C. auris.
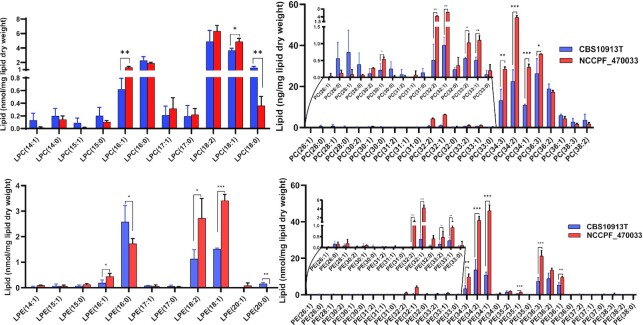
Among 149 PGL species that were detected, lipid species with 34C and 36C containing 1 to 3 double bonds in their FA chains were most abundant in NCCPF_470033 isolate. For example, lipid species 34:3, 34:2, 34:1, 36:3, 36:2, 36:1 were abundant in all PC, PE, PI, PS, PG and PA classes (Figs. 3 and 4). Majority of mono- and poly- unsaturated lipid species were present in high abundance in NCCPF_470033 strain. For example, PC (30:1, 32:2, 32:1, 33:2, 33:1, 34:3, 34:2, 34:1, 36:3), PE (32:2, 32:1, 33:2, 33:1, 34:3, 34:2, 34:1, 35:1, 36:3, 36:2, 36:1), PI (32:2, 32:1, 34:3, 34:2, 34:1, 36:1), PS (32:1, 34:1), PG (34:2, 34:1) and PA (34:3, 34:1), were in higher amounts in NCCPF_470033 as compared to CBS10913T strain (Figs. 3 and 4). Among the lysophosphoglycerides (lyso-PGL), LPC (16:1, 18:1) and LPE (16:1, 18:2, 18:1) were higher while LPC (18:0) and LPE (16:0) were lower in NCCPF_470033. (Fig. 3).
Figure 3.
Comparison of LPC, PC, LPE and PE molecular lipid species compositions between CBS10913T and NCCPF_470033 strains. The PGL species are represented as ‘total number of carbons in the acyl chains: total number of carbon-carbon double bonds in the acyl chains’. Data represents nmol per mg lipid dry weight and can be found in supplementary sheet S2. Mean ± SEM of 3 replicates is plotted. The P-values of < 0.05, <0.01 and < 0.001 are represented by *, ** and ***, respectively.
Figure 4.
Comparison of PI, PS, PG, and PA molecular lipid species compositions between CBS10913T and NCCPF_470033 strains. The PGL species are represented as ‘total number of carbons in the acyl chains: total number of carbon-carbon double bonds in the acyl chains’. Data represents nmol per mg lipid dry weight and can be found in supplementary sheet S2. Mean ± SEM of 3 replicates is plotted. The P-values of < 0.05, <0.01 and < 0.001 are represented by *, ** and ***, respectively.
Azole resistant C. auris present higher unsaturation index
A closer look at the PGL species revealed several interesting changes between NCCPF_470033 and CBS10913T strains. For example, the presence of odd chain fatty acids that have previously been also detected in other Candida species and yeasts (Singh and Prasad 2011; Kondo et al. 2014). Notably, the content of odd chain FA containing PGLs was higher in NCCPF_470033 as compared to CBS10913T strain (Fig. 5A). Although the levels of saturated PGL species did not change, the levels of unsaturated PGLs were higher in NCCPF_470033 compared to CBS10913T strain, resulting in a higher unsaturation index in NCCPF_470033 (Fig. 5B–D). The lipid ratios of PC(36:3/36:1), PE(32:2/32:1) and PE(36:3/36:1) were higher, while the ratios PE(34:2/34:1) and PE(36:2/36:1) were lower in NCCPF_470033 compared to CBS10913T strain (Fig. 5E). The changes in lipid ratios and odd chain FAs are good predictors of alteration in the membrane lipid environment (Singh and Prasad 2011).
Figure 5.
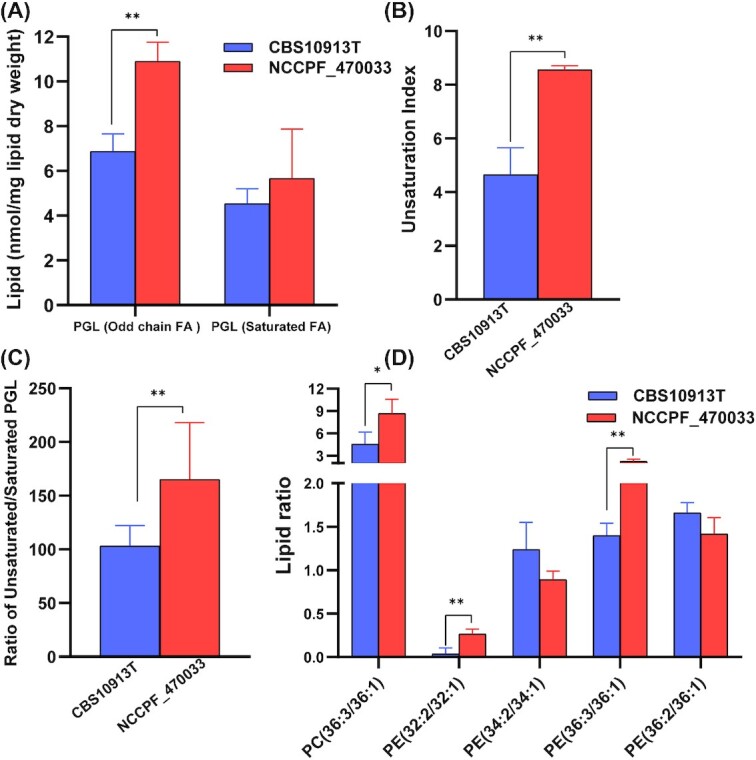
Alterations in PGL species levels of C. auris CBS10913T and NCCPF_470033 strains. (A), Total levels of odd-chain fatty acyl containing PGLs and saturated fatty acyl containing PGLs. (B), The degree of unsaturation represented as unsaturation index. The unsaturation index was calculated using the following formula: UI = [(1 × % monoene-PGL) + (2 × % diene-PGL) + (3 × % triene-PGL) + (4 × % tetraene-PGL) + (5 × % pentaene-PGL) + (6 × % hexaene-PGL)]/100. (C), Ratio of unsaturated and saturated PGLs. (D), Ratios of select PC and PE species levels. Mean ± SEM are shown (n = 3). The P-values of < 0.05, <0.01 and < 0.001 are represented by *, ** and ***, respectively.
Independent reports from others and us show that the metabolic pathways, including lipid metabolism, involved in the development of drug resistance, are often conserved in pathogenic fungi (Mukhopadhyay, Kohli and Prasad 2002; Singh and Prasad 2011; Singh, Mahto and Prasad 2013; Sharma et al. 2014; Gao et al. 2018). In this context, the available data provide credible evidence to point that imbalance in lipid homeostasis influence drug susceptibility of fungal cells (Supplementary Table S1). Notable compositional changes were recorded in earlier published comparative lipidomics between isogenic matched pair of AS/AR (azole susceptible/azole-resistant) isolates of C. albicans (Singh and Prasad 2011). In Gu4/Gu5, G2/G5, YOI-16/YOI-64 isolates representing AS (azole sensitive/AR (azole resistant) isogenic pairs of C. albicans hospital isolates showed consistent increase in odd chain FA containing PGLs and unsaturation index in AR G5, Gu5, and YOI-64 isolates as compared to susceptible Gu4, G2 and YOI-16, respectively (Singh and Prasad 2011; Singh, Mahto and Prasad 2013). The odd chain FAs have a better advantage in influencing the membrane fluidity as compared to the polyunsaturated FAs (Jenkins, West and Koulman 2015), probably due to its low melting temperature (Ibarguren, López and Escribá 2014; Beppu et al. 2017). The compromised PGL biosynthesis with resulting changes in membrane fluidity affects was shown to affect the drug diffusion, accumulation and drug susceptibility of C. albicans cells highlighting a strong link between PGL homeostasis and azole resistance (Singh and Prasad 2011). The raised unsaturated lipids content in the present study is another common compositional change between resistant C. auris (present study) and C. albicans cells (Singh and Prasad 2011).
NCCPF_470033 shows variations in DAG and TAG metabolism
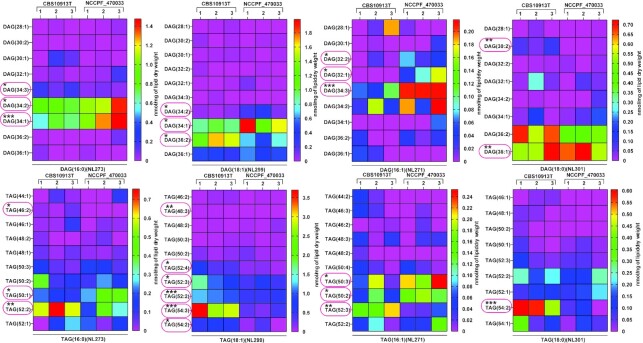
Among the neutral lipids, of a majority of DAG species that showed statistically significant changes, 16:1-DAG (32:2, 32:1, 34:3), 16:0-DAG (34:2, 34:1) and 18:1-DAG (34:2, 34:1) species were abundant in NCCPF_470033 compared to CBS10913T strain (Fig. 6). In contrast, of a majority of TAG species that showed statistically significant changes, 16:1-TAG (52:3), 16:0-TAG (46:2, 52:2), 18:1-TAG (48:3, 52:4, 52:3, 52:2, 54:3, 52:2) and 18:0-TAG (54:2) were present in much low in NCCPF_470033 compared to CBS10913T strain (Fig. 6).
Figure 6.
Comparison of DAG and TAG molecular lipid species compositions in CBS10913T and NCCPF_470033 strains. The DAG and TAG molecular species were analyzed as described earlier (Singh and Prasad 2011), also see Figure 2). DAG and TAG species are represented ‘total number of carbons in the acyl chains: total number of carbon-carbon double bonds in the acyl chains’. DAG and TAG species boxed in red represents statistically significant changes. Data represents nmol per mg lipid dry weight and can be found in supplementary sheet S2. All the 3 replicates are plotted for each strain. The P-values of < 0.05, <0.01 and < 0.001 are represented by *, ** and ***, respectively.
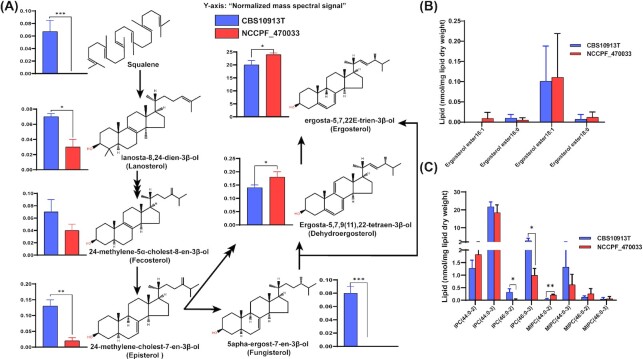
Resistant C. auris possess a higher level of free ergosterol
Sterols and SLs are the other two essential components of the membranes that directly influence membrane fluidity (Dufourc 2008; Guan et al. 2009). Azole resistance in C. auris is caused by alteration in the ergosterol pathway and ergosterol content (Kean and Ramage 2019; Zamith-Miranda et al. 2019). We examined ergosterol and its intermediates by employing GC-MS and identified seven key intermediates of ergosterol biosynthesis pathway. Upon analyzing the free ergosterol biosynthesis intermediates, we observed that, most were significantly lower in NCCPF_470033 as compared to CBS10913T. Remarkably both fungisterol and squalene were below the detection limit and showed a drastic reduction in NCCPF_470033 (Fig. 7A). The end products ergostatetraenol and ergosterol levels, however, were significantly higher in NCCPF_470033 as compared to CBS10913T strain. In contrast, the levels of ergosteryl esters remained unchanged between the two isolates (Fig. 7B). similarly, total ergosterol was found higher in azole resistant C. auris isolate (Zamith-Miranda et al. 2019). The higher ergosterol content in NCCPF_470033 well correlated with observed higher resistance towards azoles of NCCPF_470033 isolate (Fig. 7A). The lowering of ergosterol under hypoxic C. albicans cells reported recently corroborates earlier studies. It highlights its role in governing FLC resistance (Burgain et al. 2020).
Figure 7.
Comparison of sterols and sphingolipid compositions in CBS10913T and NCCPF_470033 strains. (A), Analysis of free sterol species. (B), Analysis of steryl esters. (C), Analysis of IPC's and MIPC's. IPC and MIPC species are represented as ‘total number of carbons in the sphingoid base and acyl chains: total number of carbon-carbon double bonds in the sphingoid base and acyl chains- number of hydroxyl groups present in the sphingoid base and acyl chains’. Data in (A) is represented as the normalized mass spectral signal. Data in (B) and (C) are represented as nmol per mg lipid dry weight and can be found in supplementary sheet S2. Mean ± SEM of 3 replicates is plotted. The P-values of < 0.05, <0.01 and < 0.001 are represented by *, ** and ***, respectively.
Few complex SL species show alteration in NCCPF_470033
Although the detailed analysis of SLs requires different protocol of extraction, the method employed in the present study let identification of some of the SL structures. Our analysis revealed that the levels of IPC (44:0–2), IPC (46:0–3) and MIPC (44:0–2) species were significantly lowered in NCCPF_47033 compared to CBS10913T strain (Fig. 7C). Some of IPC structures detected did show variation between CBS10913T and NCCPF_470033 strains, and whether these changes are related to drug susceptibilities will require further studies. A detailed and targeted sphingolipidomic study is required to understand the relevance of SLs in drug resistance in C. auris.
PCA analysis reveals the distinctness of the susceptible and resistant isolate
The comparative lipidome data described above hints towards a massive remodeling of lipid species in NCCPF_470033 when compared to the CBS10913T strain. We performed PCA analysis to further examine the statistical significance of remodeling of lipids associated with high azole resistance. PCA analysis allows the extraction and visualization of statistically significant variations from larger data sets. The extracted data are described by the principal components which account for the possible variations within the data. PCA was performed, using the lipid species data in nmol/mg lipid weight, between NCCPF_470033 and CBS10913T strains to highlight the statistically significant SL variations amongst them. The PCA analysis allowed the extraction of 5 principal components, of which the principal component 1, 2 and 3 accounted for ∼83% variance in the data sets and were plotted (Fig. 8). Also, the associated loading values of each lipid species are decisive in the allocation of each principal component.
Figure 8.
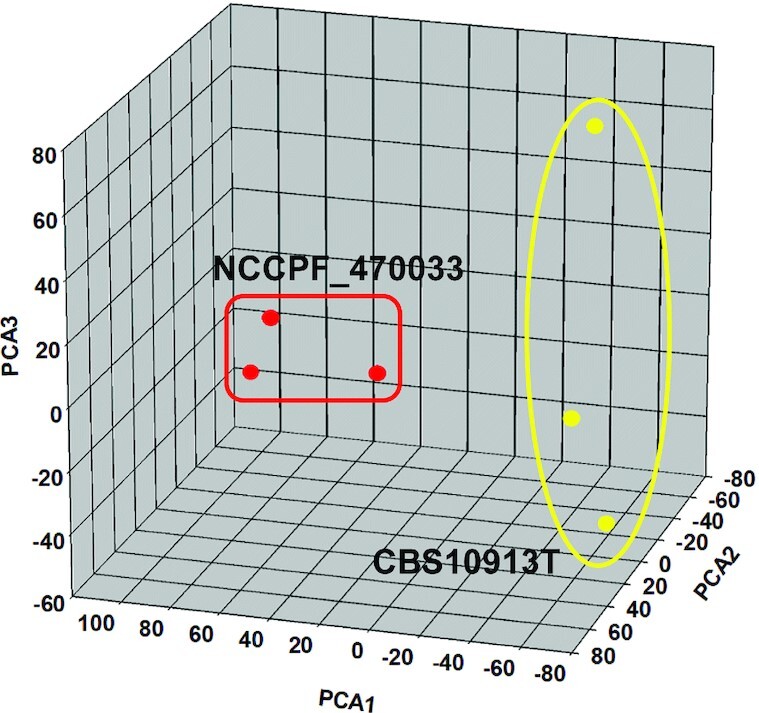
PCA analysis of lipid species compositions of CBS10913T and NCCPF_470033 strains of C. auris. PCA analysis was performed using the XLSTAT software described in methods. The scores of the top 3 principal components (factors) are plotted, demonstrating more than 83% of the variance in the data sets. The principal component 1 (factor 1), principal component 2 (factor 2) and principal component 3 (factor 3) contribute 43.33%, 24.5% and 15.5% variances respectively. A blind analysis was performed using 3 replicate data sets of each strain (Supplementary sheet S2). A list of factors loading values associated with each principal component is shown in supplementary sheet S4.
The plot of principal components 1 and 2 exhibited segregation of CBS10913T and NCCPF_470033 strains from each other (Fig. 8). An inspection of the loading values linked to principal component 1 confirmed that the lipid species with less abundance (like TAG(52:3)-16:1, TAG(54:2)-18:0, TAG(54:3)-18:1) and those with high abundance (like DAG(32:1)-16:1 and DAG(34:1)-18:1) in azole resistant NCCPF_470033 strain (compared to CBS10913T), are key to the separation of CBS10913T and NCCPF_470033 strains on principal component 1 axis. Analysis of loading values linked to principal component 2 reveals that the lipid species with less abundance (like TAG(54:2)-18:0, TAG(52:2)-16:0, TAG(52:3)-16:1) and those with high abundance (like DAG(34:1)-18:1, DAG(34:3)-16:1, DAG(32:1)-16:1) in NCCPF_470033 (compared to CBS10913T), are key to separation of CBS10913T and NCCPF_470033 strains on principal component 2 axis. Similar variations were also observed across principal component 3 axis among the DAG and TAG species
However, the PCA analysis using the complete lipid species data set could not highlight the significant changes in the phospholipid group. To resolve this issue, we performed yet another PCA analysis using the lipid species data as mole% (% of total PGL + SL, supplementary sheet S3). In these analyses, five principal components were extracted, where the principal component 1 and 2 accounted for more than 95% variation in the data sets (Fig. S2a, Supporting Information). The loading values of principal component 1 show that PC (36:3, 36:2, 36:1, 38:3), PA (36:2, 36:1), PI (36:3), IPC (44:0–3), LPE (16:0) are deficient, while PE (32:1, 34:1, 34:2, 34:3, 36:3), PC (32:1, 32:2, 34:1, 34:2) are abundant, in NCCPF_470033 strain and important for segregation of CBS10913T and NCCPF_470033 strains on principal component 1 axis (Fig. S2a and b, Supporting Information). The loading values of principal component 2 show that the mono-unsaturated lipid species like PC (28:1, 36:1), PI (36:1), IPC (44:0–3), LPC (16:0) are deficient. In contrast, poly-unsaturated lipid species like PE (34:2, 34:3, 36:3), PC (34:2, 34:3) are abundant, in NCCPF_470033 strain and important for segregation of CBS10913T and NCCPF_470033 strains on principal component 2 axis (Fig. S2a and b, Supporting Information). Together, these PCA analyses highlight the specific subset of lipid species associated specifically with the azole resistant C. auris.
A comparative PCA analysis between the lipid profiles of C. auris strains (this study) with those of other Candida species (published previously, (Singh et al. 2010) shows a distinct lipid imprint of C. auris (Fig. S2c and d, Supporting Information). Further, using the PCA analysis, we could also highlight similarities and differences in lipid profiles of C. auris strains and the previously published lipidomes of clinical isolates of C. albicans (Singh and Prasad 2011; Singh, Yadav and Prasad 2012; Singh, Mahto and Prasad 2013) (Supplementary Fig. S2e and S2f).
Although the PCA analyses did highlight certain lipid species that show significant changes between data sets, however, lipid changes consistent with drug resistance are difficult to often understand. For simplification, we have summarized the lipid variations reported in earlier published studies involving AS/AR isolates of C. albicans and C. auris and the present study (Table S1, Supporting Information). A comparison of these data sets shows that the lipid changes occurring between different AS/AR isolates are quite divergent and could impact drug susceptibilities in different ways, and demand further studies for better understanding.
CONCLUSION
Overall, in this study we were able to: (i) quantify major lipid groups and determine molecular lipid species compositions; (ii) highlight specific changes among the lipid species associated with azole resistance in C. auris; (iii) underline the commonalities of lipid compositional changes between AR C. auris and other Candida spp.; (iv) identify that specific lipid species variations could be a feature to distinguish C. auris strains from the other species of Candida. In future, we aim to expand our spectrum of analysis by including several susceptible and resistant isolates that are resistant to fluconazole and show collateral resistance to other class of drugs to pave way for better insight into the relevance of lipids in drug resistance in this important pathogenic yeast.
AUTHORSHIP
Lipid experimentation and data analysis: GS, MK and AS; Strain curation and drug susceptibility: SK, MK, SMR and AC; Manuscript writing and review editing: MK, AS, AC, NAG and RP; Funding: AS, NAG and RP; Conceptualization: AS and RP. All authors read and approved the final manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
RP acknowledges support from the Indian Council of Medical Research, ICMR (AMR/149 (2)-2018-ECD-II). AS thanks support from ICMR (No. 52/08/2019-BIO/BMS), DST-PURSE program (SR/PURSE Phase 2/29(C)) and the University of Lucknow. AS recognizes the support of Amity University, Haryana, during a mini-sabbatical stay. NAG acknowledges ICGEB, New Delhi, and Department of Biotechnology (DBT), Government of India for financial support (Grant No.: BT/PB/Centre/03/ICGEB/2011-PhaseII). The lipid analyses described in this work were performed in parts at the Kansas Lipidomics Research Center Analytical Laboratory, Kansas State University, USA (funded by NSF grants EPS 0236913, MCB 1413036, MCB 0920663, DBI 0521587, DBI1228622 and K-INBRE of NIH P20GM103418) and Amity Lipidomics Research Facility (ALRF), Amity University Haryana, India. MK and SK acknowledge the Department of Biotechnology (DBT) and the Council of Scientific and Industrial Research (CSIR) for providing fellowship support.
Contributor Information
Garima Shahi, Amity Institute of Integrative Science and Health and Amity Institute of Biotechnology, Amity University Gurugram,, Haryana, 122413, India.
Mohit Kumar, Amity Institute of Integrative Science and Health and Amity Institute of Biotechnology, Amity University Gurugram,, Haryana, 122413, India; Yeast Biofuel Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India.
Sonam Kumari, Yeast Biofuel Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India.
Shivaprakash M Rudramurthy, Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research,, Chandigarh, 160012, India.
Arunaloke Chakrabarti, Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research,, Chandigarh, 160012, India.
Naseem A Gaur, Yeast Biofuel Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India.
Ashutosh Singh, Department of Biochemistry, University of Lucknow, Lucknow, Uttar Pradesh, 226007, India.
Rajendra Prasad, Amity Institute of Integrative Science and Health and Amity Institute of Biotechnology, Amity University Gurugram,, Haryana, 122413, India.
Conflict of interest
None declared.
REFERENCES
- Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol. 2005;3:547–56. [DOI] [PubMed] [Google Scholar]
- Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–6. [DOI] [PubMed] [Google Scholar]
- Beppu F, Yasuda K, Okada Aet al. . Comparison of the Distribution of Unsaturated Fatty Acids at the Sn-2 Position of Phospholipids and Triacylglycerols in Marine Fishes and Mammals. J Oleo Sci. 2017;66:1217–27. [DOI] [PubMed] [Google Scholar]
- Bowden JA, Heckert A, Ulmer CZet al. . Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J Lipid Res. 2017;58:2275–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NARet al. . Hidden Killers: Human Fungal Infections. Sci Transl Med. 2012;4:165rv13. [DOI] [PubMed] [Google Scholar]
- Burgain A, Tebbji F, Khemiri Iet al. . Metabolic Reprogramming in the Opportunistic Yeast Candida albicans in Response to Hypoxia. mSphere. 2020;5, DOI: 10.1128/mSphere.00913-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Sharma C, Meis JF. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. Hogan DA (ed). PLoS Pathog. 2017;13:e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourc EJ. Sterols and membrane dynamics. J Chem Biol. 2008;1:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairlamb AH, Gow NA, Matthews KRet al. . Stop neglecting fungi. Nat Microbiol. 2017;2:17120. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gao J, Wang H, Li Zet al. . Candida albicans gains azole resistance by altering sphingolipid composition. Nat Commun. 2018;9:4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves S, Silva PM, Felício MRet al. . Psd1 Effects on Candida albicans Planktonic Cells and Biofilms. Front Cell Infect Microbiol. 2017;7:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler Het al. . Functional Interactions between Sphingolipids and Sterols in Biological Membranes Regulating Cell Physiology. Mol Biol Cell. 2009;20:2083–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Healey KR, Shor Eet al. . Novel FKS1 and FKS2 modifications in a high-level echinocandin resistant clinical isolate of Candidaglabrata. Emer Microb Infect. 2019;8:1619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarguren M, López DJ, Escribá PV. The effect of natural and synthetic fatty acids on membrane structure, microdomain organization, cellular functions and human health. Biochim Biophys Acta BBA - Biomembr. 2014;1838:1518–28. [DOI] [PubMed] [Google Scholar]
- Jenkins B, West JA, Koulman A. A Review of Odd-Chain Fatty Acid Metabolism and the Role of Pentadecanoic Acid (C15:0) and Heptadecanoic Acid (C17:0) in Health and Disease. Molecules. 2015;20:2425–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria S, Singh PK, Sharma Cet al. . Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. Warnock DW (ed.). J Clin Microbiol. 2015;53:1823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean R, Ramage G. Combined Antifungal Resistance and Biofilm Tolerance: the Global Threat of Candida auris. mSphere. 2019;4, DOI: 10.1128/mSphere.00458-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal NK, Sarkar P, Gaur NAet al. . Phosphatidylserine decarboxylase governs plasma membrane fluidity and impacts drug susceptibilities of Candida albicans cells. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2018;1860:2308–19. [DOI] [PubMed] [Google Scholar]
- Kim SH, Iyer KR, Pardeshi Let al. . Genetic Analysis of Candidaauris Implicates Hsp90 in Morphogenesis and Azole Tolerance and Cdr1 in Azole Resistance. Kronstad JW (ed). mBio. 2019;10:e02529–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Ohno Y, Yamagata Met al. . Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat Commun. 2014;5:1–13. [DOI] [PubMed] [Google Scholar]
- Kumari S, Kumar M, Khandelwal NKet al. . ABC transportome inventory of human pathogenic yeast Candida glabrata: Phylogenetic and expression analysis. PLoS One. 2018;13:e0202993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattif AA, Mukherjee PK, Chandra Jet al. . Lipidomics of Candida albicans biofilms reveals phase-dependent production of phospholipid molecular classes and role for lipid rafts in biofilm formation. Microbiology. 2011;157:3232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Etienne KA, Vallabhaneni Set al. . Simultaneous Emergence of Multidrug-Resistant Candidaauris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. CLINID. 2017;64:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone SA, Ahmad A.. Candida auris —the growing menace to global health. Mycoses. 2019;62:620–37. [DOI] [PubMed] [Google Scholar]
- Lupetti A, Danesi R, Campa Met al. . Molecular basis of resistance to azole antifungals. Trends Mol Med. 2002;8:76–81. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay K, Kohli A, Prasad R. Drug Susceptibilities of Yeast Cells Are Affected by Membrane Lipid Composition. Antimicrob Agents Chemother. 2002;46:3695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Boyken LB, Hollis RJet al. . Validation of 24-Hour Fluconazole MIC Readings versus the CLSI 48-Hour Broth Microdilution Reference Method: Results from a Global Candida Antifungal Surveillance Program. J Clin Microbiol. 2008;46:3585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Singh A. Lipids of Candida albicans and their role in multidrug resistance. Curr Genet. 2013;59:243–50. [DOI] [PubMed] [Google Scholar]
- Prasad T, Saini P, Gaur NAet al. . Functional Analysis of CaIPT1, a Sphingolipid Biosynthetic Gene Involved in Multidrug Resistance and Morphogenesis of Candida albicans. Antimicrob Agents Chemother. 2005;49:3442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak JM, Doorley LA, Nishimoto ATet al. . Abrogation of Triazole Resistance upon Deletion of CDR1 in a Clinical Isolate of Candida auris. Antimicrob Agents Chemother. 2019;63, DOI: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Makimura K, Hasumi Yet al. . Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–4. [DOI] [PubMed] [Google Scholar]
- Sharma M, Dhamgaye S, Singh Aet al. . Lipidome analysis reveals antifungal polyphenol curcumin affects membrane lipid homeostasis. Front Biosci (Elite Ed). 2012;4:1195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Alfatah, Bari VKet al. . Sphingolipid Biosynthetic Pathway Genes FEN1 and SUR4 Modulate Amphotericin B Resistance. Antimicrob Agents Chemother. 2014;58:2409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Perlin DS.. Coping with stress and the emergence of multidrug resistance in fungi. PLoS Pathog. 2015;11:e1004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Del Poeta M. Sphingolipidomics: An Important Mechanistic Tool for Studying Fungal Pathogens. Front Microbiol. 2016;7, DOI: 10.3389/fmicb.2016.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, MacKenzie A, Girnun Get al. . Analysis of sphingolipids, sterols, and phospholipids in human pathogenic Cryptococcus strains. J Lipid Res. 2017;58:2017–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Mahto KK, Prasad R. Lipidomics and in Vitro Azole Resistance in Candida albicans. OMICS: J Int Biol. 2013;17:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Prasad R. Comparative Lipidomics of Azole Sensitive and Resistant Clinical Isolates of Candida albicans Reveals Unexpected Diversity in Molecular Lipid Imprints. PLoS One. 2011;6:e19266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Prasad T, Kapoor Ket al. . Phospholipidome of Candida: Each Species of Candida Has Distinctive Phospholipid Molecular Species. OMICS: J Int Biol. 2010;14:665–77. [DOI] [PubMed] [Google Scholar]
- Singh A, Yadav V, Prasad R. Comparative Lipidomics in Clinical Isolates of Candida albicans Reveal Crosstalk between Mitochondria, Cell Wall Integrity and Azole Resistance. PLoS One. 2012;7:e39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasi M, Khandelwal NK, Moorhouse AJet al. . ABC Transporter Genes Show Upregulated Expression in Drug-Resistant Clinical Isolates of Candida auris: A Genome-Wide Characterization of ATP-Binding Cassette (ABC) Transporter Genes. Front Microbiol. 2019;10:1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamith-Miranda D, Heyman HM, Cleare LGet al. . Multi-omics Signature of Candidaauris, an Emerging and Multidrug-Resistant Pathogen. Elias J (ed). mSystems. 2019;4:e00257–19., /msystems/4/4/mSys.00257-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.