Figure 2.
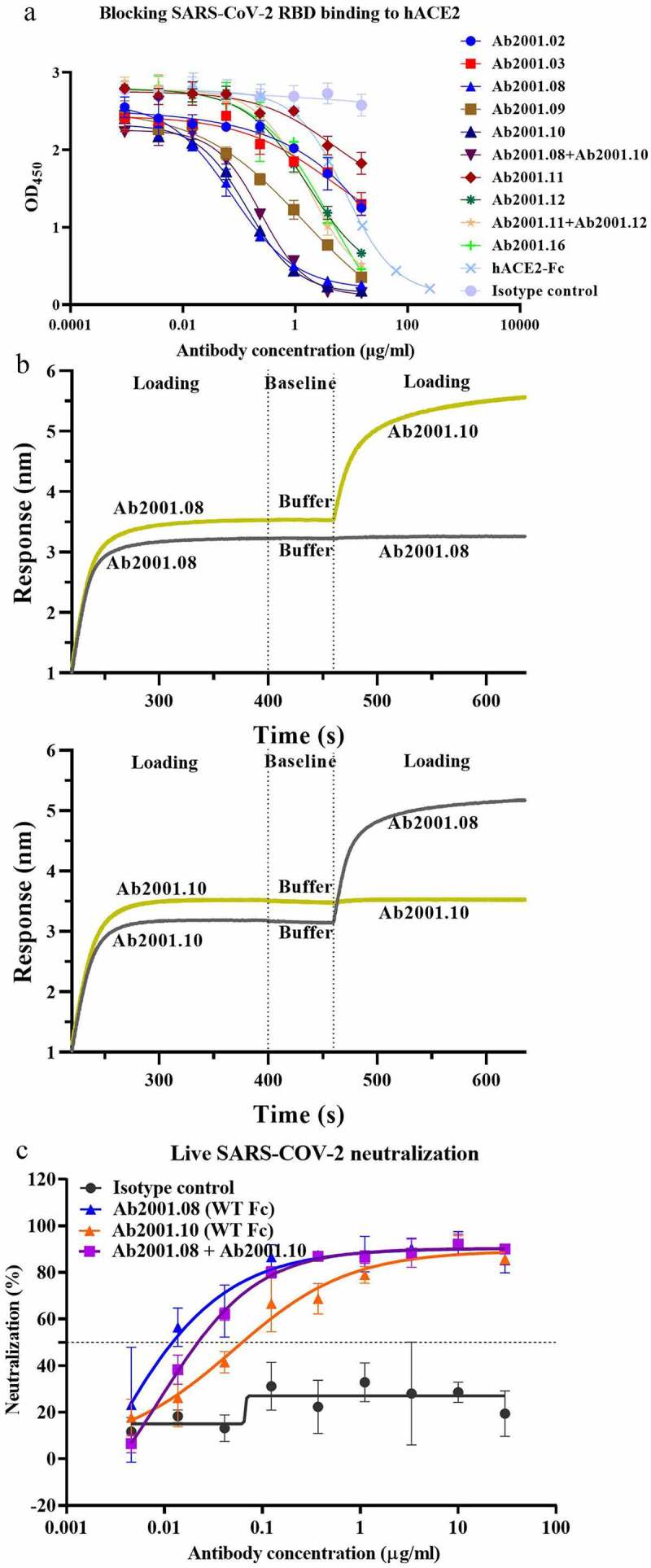
Characterization of potential blocking antibodies. (a) Blocking assay was performed by immobilizing 1 µg/ml hACE2 on a plate. Serially diluted antibodies and biotinylated SARS-CoV-2 RBD protein were added for competitive binding to hACE2. IC50 values were calculated with Prism V8.0 software using a four-parameter logistic curve fitting approach. (b) Epitope binning was carried out by BLI. Biotinylated SARS-CoV-2 RBD was immobilized onto the SA sensor, and a high concentration of the primary antibody was used to saturate its own binding site. Subsequently, a second antibody was applied to compete for the binding site on the SARS-CoV-2 RBD protein. Data were analyzed with Octet Data Analysis HT 11.0 software. (c) Neutralization activities of Ab2001.08 and Ab2001.10 were assessed by live virus assay. Live SARS-CoV-2 and serially diluted (3-fold) antibodies were added to VERO E6 cells. The PRNT50 values were determined by plotting the plaque number (neutralization percentage) against the log antibody concentration in Prism V8.0 software
