Figure 3. Cryo-EM density map and filament model for the N. gonorrhoeae T4P.
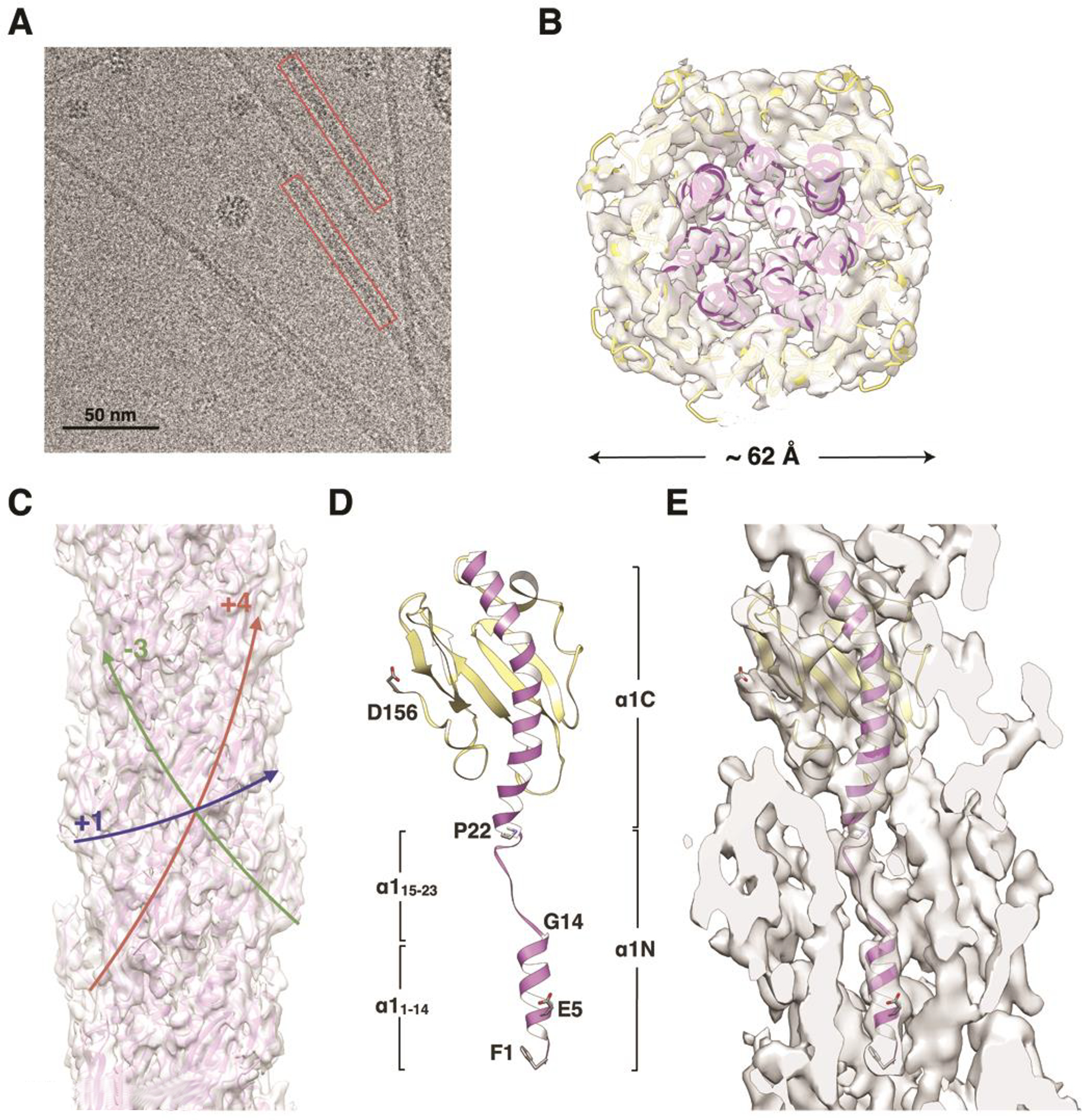
(A) Cryo-electron micrograph of N. gonorrhoeae T4P. (B) Slice of the Ng pilus reconstruction and filament model viewed along the filament axis. (C) Side view of the PaK pilus reconstruction and filament model. Colored arrows indicate the paths of the (+)1-, (−)3- and (+)4-start helices. (D) Ribbon representation of the Ng PilE model generated by fitting the protein into the cryo-EM density in three segments: the globular domain and α1:1–14 were fit separately as rigid bodies from the PilE crystal structure 2HI2, and α1:15–23 was modeled in an extended conformation. α1 is purple and the remainder of the globular domain is yellow. This subunit model was used to generate the Ng pilus filament model. (E) Slice through the cryo-EM density map showing a single PilE subunit. See also Figure S1B, Movie S2.
