Figure 6. Conserved residues in the Type IV pilin α1 are critical for pilus assembly and functions.
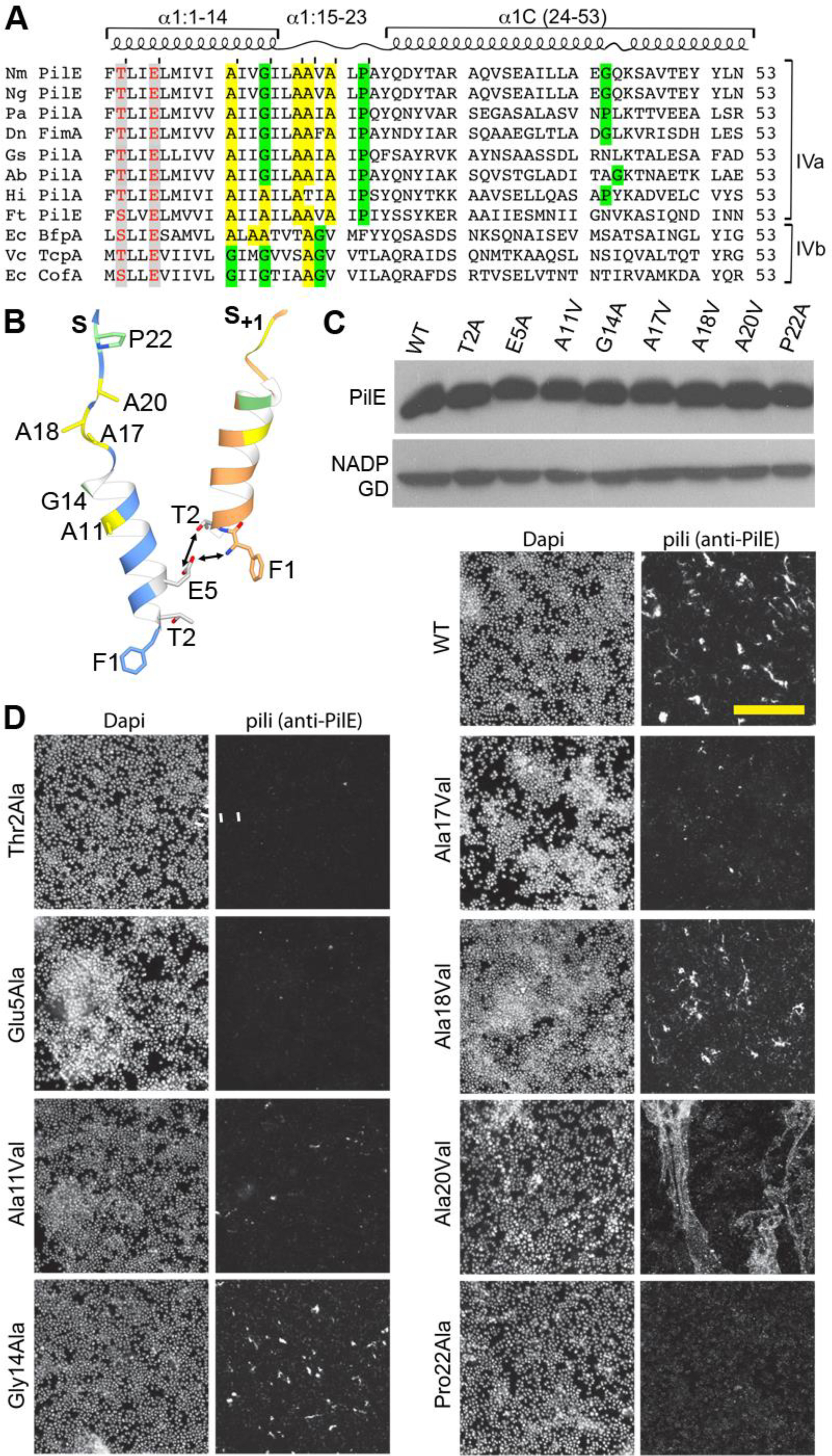
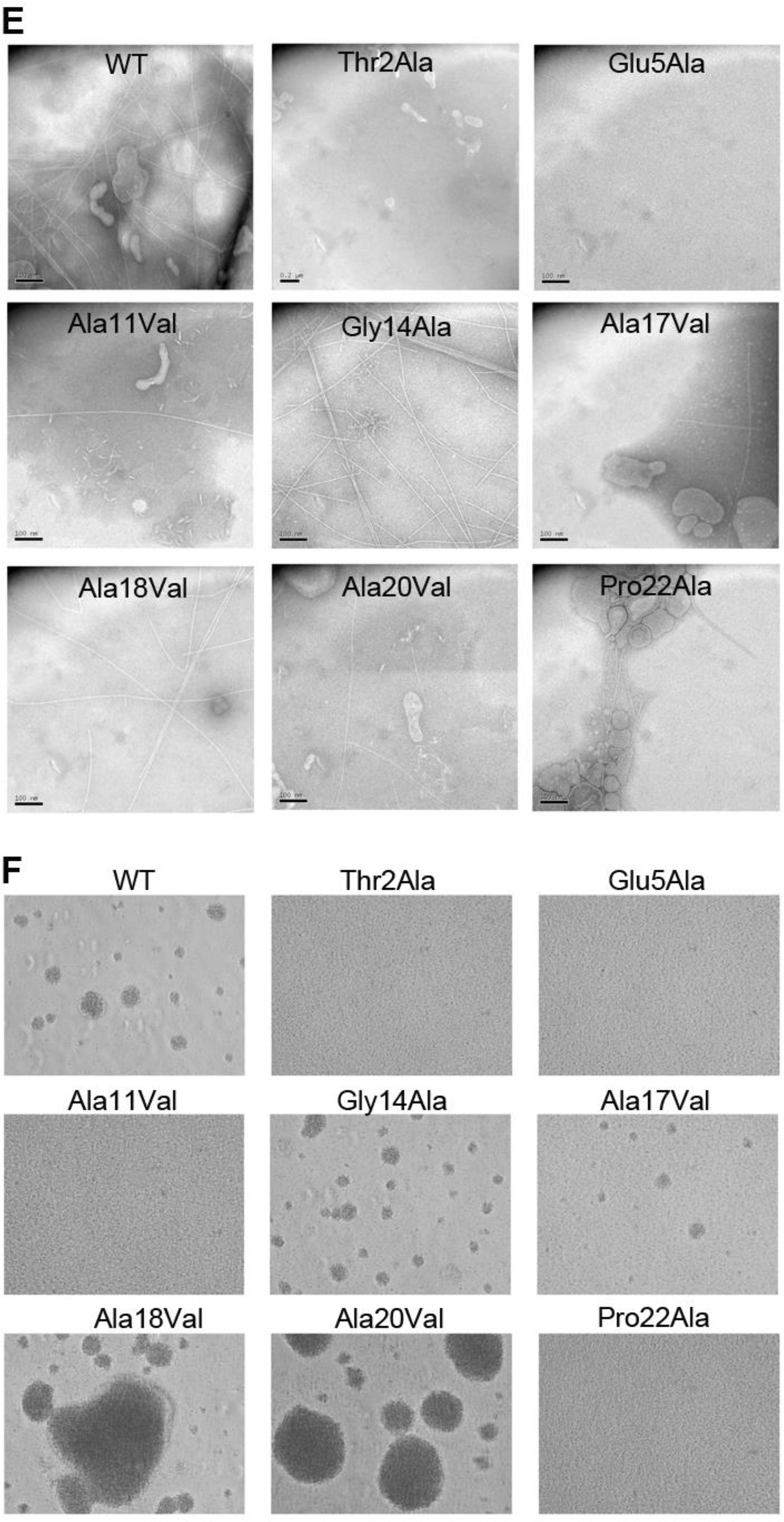
(A) Amino acid sequence alignment of α1 (residues 1–53) from Type IVa and Type IVb pilins. The α-helical portions of this segment are indicated based on the cryo-EM density maps for Nm, Ng and PaK T4P, and the melted region is boxed. Thr/Ser2 and Glu5 are highlighted in grey with red text. Conserved alanines within or near the melted region are highlighted yellow and the conserved helix-breaking residues proline and glycine are highlighted green. Nm PilE, N. meningitidis PilE (GenBank accession number WP_014573675); Ng PilE, N. gonorrhoeae PilE (for 2HI2, P02974); PaK PilA, P. aeruginosa PilA (P02973); Dn FimA, Dichelobacter nodosus (X52403), Gs PilA, Geobacter sulfurreducins PilA (2M7G_A); Ab PilA, Acinetobacter baumannii ACICU (ACC58690); Hi, non-typeable Haemophilus influenzae (AAX87353); Ft PilE1, Francisella tularensis Schu S4 PilE1 (CAG45522); Ec BfpA, enteropathogenic Escherichia coli BfpA (Z68186); Vc TcpA, Vibrio cholerae TcpA (ABQ19609); Ec CofA, enterotoxigenic E. coli (CofA, BAA07174) (B) Close-up of the N-terminus of two PilE subunits in the (+)1 helix of Nm T4P model (Kolappan et al., 2016) showing the melted α1:15–23 segment and the interactions between Glu5 of subunit S and Phe1 and Thr2 in subunit S+1 in the (+)1 helix. (C) Immunoblot of whole cell lysates of N. meningitidis SB-Kn (WT) and Sb-Kn pilE mutants probed with antibodies against PilE and a cytosolic marker protein, NADP glutamate dehydrogenase. (D) Immunofluorescent labeling of N. meningitidis WT and pilE mutant strains stained with DAPI to visualize the bacteria and anti-PilE and goat anti-mouse Alexa 488 antibody to visualize the pili. The scale bar is 20 μm. (E) Aggregation of N. meningitidis WT and pilE mutant strains imaged on an inverted microscope after overnight growth and resuspension in fetal bovine serum. (F) TEM imaging of pili sheared from N. meningitidis WT and pilE mutant strains.
