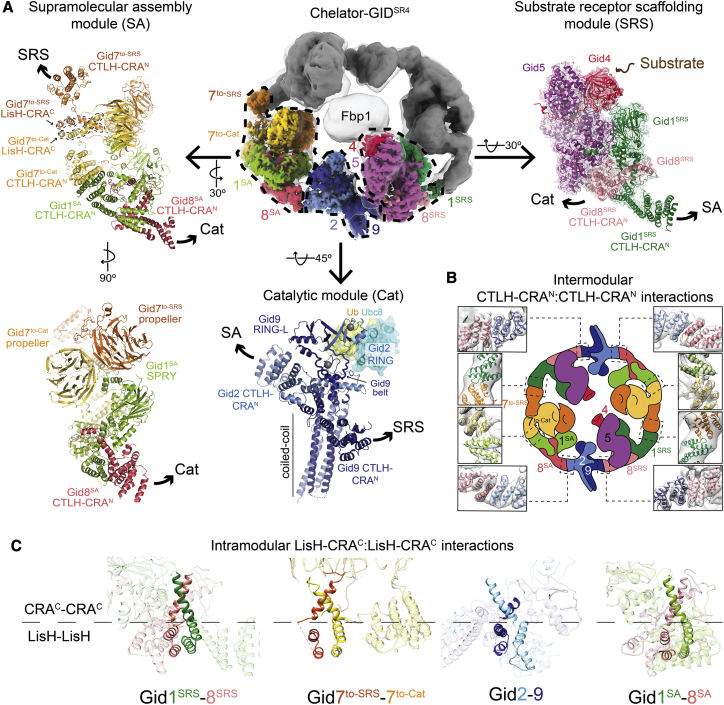
Figure 3.
High-resolution details of Chelator-GIDSR4 modular assembly
(A) Focused refined maps of the substrate receptor scaffolding (SRS), catalytic (Cat), and supramolecular assembly (SA) modules, colored according to subunit identity, fit in half of the overall map of Fbp1-bound Chelator-GIDSR4 (top center). The GIDSR4 structure (PDB: 6SWY) fits the SRS module (Gid1SRS, dark green; Gid8SRS, salmon; Gid5, purple; Gid4, red). A brown arrow points to Gid4’s substrate binding site (top right). The Cat module comprises Gid2 (sky blue) and Gid9 (navy). Zinc ions are shown as gray spheres. Ubc8~Ub was modeled by aligning Gid2 RING with an E2~Ub-bound RING structure (PDB: 5H7S). The SA module comprises Gid1SA (green), Gid8SA (pink) and 2 Gid7 protomers, Gid7to-Cat (yellow), and Gid7to-SRS (orange) facing the Cat or SRS module, respectively. Superscript text refers to a module for a given Gid1 or Gid8 protomer. Arrows point to connected modules.
(B) Cartoon of Chelator-GIDSR4 with close ups of intermodule CTLH-CRAN:CTLH-CRAN interactions fit into the map of Chelator-GIDSR4 (gray).
(C) Intramodule LisH-CRAC:LisH-CRAC (solid ribbon) interactions in Chelator-GIDSR4.
See also Figures S3 and S4 and Table S1.