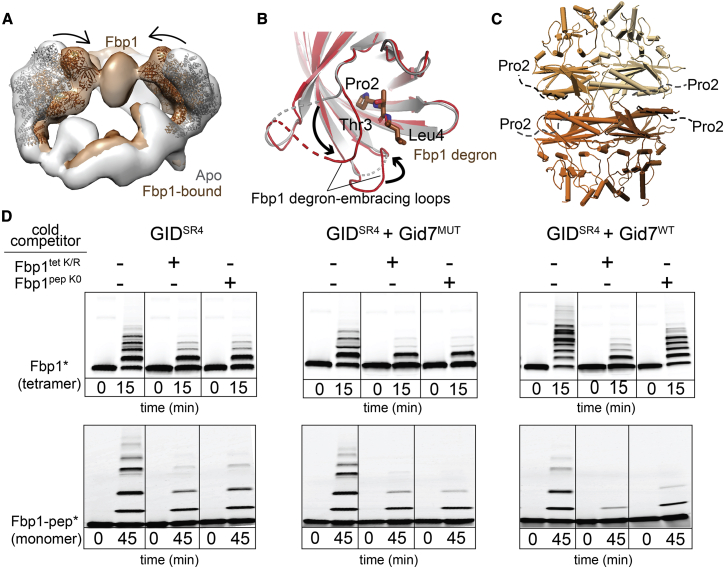
Figure 4.
Chelator-GIDSR4 assembly specifies multivalent binding for the tetrameric Fbp1 substrate
(A) Superimposed maps of substrate-free (gray) and Fbp1-bound Chelator-GIDSR4 (brown) show relative inward movement of SRS modules (ribbon) upon substrate recruitment.
(B) Conformational differences between Gid4 in GIDSR4 (PDB: 6SWY, gray) and Fbp1-bound Chelator-GIDSR4 (red). The first three residues of Fbp1 (the Pro/N-degron) bound to Gid4 are shown as sticks.
(C) Crystal structure of the Fbp1 tetramer, with the N-terminal region (residues 2–19), including the degron not visible in the electron density, depicted as dotted lines. Fbp1 protomers are shown in various brown shades.
(D) Competitive in vitro ubiquitylation assays probing multivalent E3-substrate interactions. Chelator-GIDSR4 has two substrate binding sites and two catalytic centers, whereas two other E3 assemblies (GIDSR4 or GIDSR4 + Gid7MUT lacking the LisH-CTLH-CRA motif, Δ1–285) have only one substate binding site and one catalytic center. Substrates are oligomeric (tetrameric Fbp1) or monomeric (a peptide harboring a single acceptor Lys, Fbp1-pep) and fluorescently labeled at the C terminus (denoted by an asterisk). Competitors are oligomeric (tetrameric Fbp1tet K/R, with preferred target lysines mutated to arginines) or monomeric (lysine-less peptide, Fbp1pep K0).