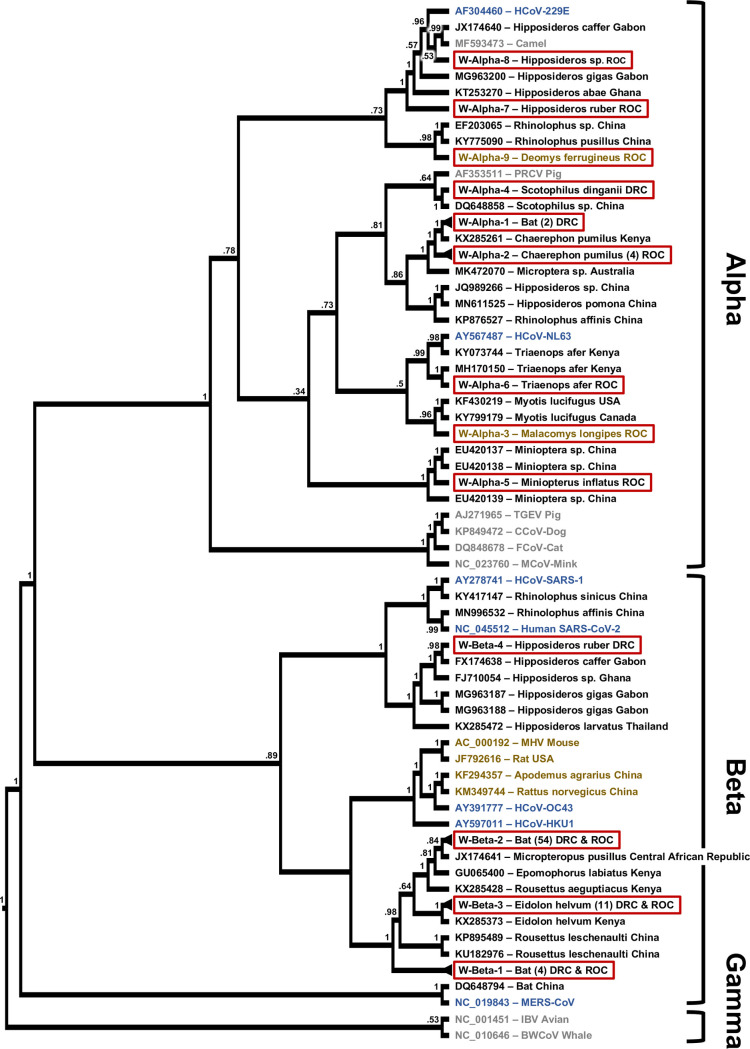
Fig 2. Phylogenetic tree based on the RdRp region targeted by the PCR by Watanabe.
Maximum likelihood phylogenetic tree of coronavirus sequences presented as a proportional cladogram, based on the RdRp region targeted by the PCR by Watanabe et. al. [41]. The tree includes the sequences detected during the project (red boxes) and indicates the number of sequences sharing more than 95% nucleotide identities in brackets. GenBank accession numbers are listed for previously published sequences, while sequences obtained during the project are identified by cluster names (compare S2 Table). Black font indicates coronavirus sequences obtained from bats, brown font indicates rodents, blue humans and gray other hosts. The host species and country of sequence origin are indicated for bats and rodents if applicable. In case of clusters W-Alpha-1 sequences were detected in Mops condylurus and Chaerephon sp., host species in cluster W-Beta-1 were Megaloglossus woermanni and Epomops franqueti and in case of cluster W-Beta-2 Micropteropus pusillus, Epomops franqueti, Rhinolophus sp., Myonycteris sp., Mops condylurus, Megaloglossus woermanni, and Eidolon helvum (compare S2 Table). Numbers at nodes indicate bootstrap support.