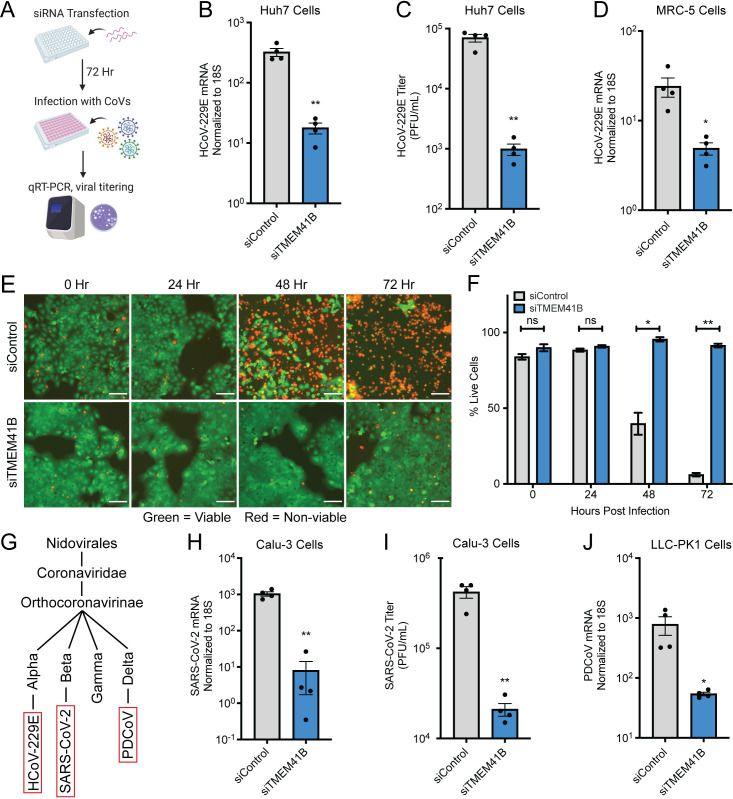
Fig 2. TMEM41B is essential for the replication of multiple coronaviruses.
(A) Experimental scheme for siRNA-mediated targeting of TMEM41B. (B) HCoV-229E RNA quantified in siRNA-transfected Huh7 cells. N = 4. (C) HCoV-229E titer measured via plaque assay after infection of siRNA-transfected Huh7 cells. Supernatant was collected for titering 48 HPI, N = 4. (D) HCoV-229E RNA quantified in siRNA-transfected MRC-5 cells. (E) Live (green) and dead (red) cell staining after control or TMEM41B knockdown at the indicated timepoints. Scale bars are 100 μm. (F) Quantification of E. N = 3 independently captured images. (G) Simplified representation of the phylogenetic relationship between HCoV-229E, SARS-CoV-2, and PDCoV. (H) SARS-CoV-2 RNA quantified in siRNA-transfected Calu-3 cells. (I) SARS-CoV-2 titer measured via plaque assay after infection of Calu-3 cells. Supernatant was collected for titering 48 HPI, N = 4. (J) PDCoV RNA quantified in siRNA-transfected LLC-PK1 cells. Cells were infected with a dilution of PDCoV such that viral replication was detectable by qRT-PCR. Unless otherwise specified, MOI = 0.01 for all experiments and samples were collected at 24 HPI. Error bars represent standard error measurement. Significance values were determined using a two-tailed, unpaired, Student’s t-test. *P<0.05, **P<0.001, ns = not significant. Data are representative of at least two independently conducted experiments.