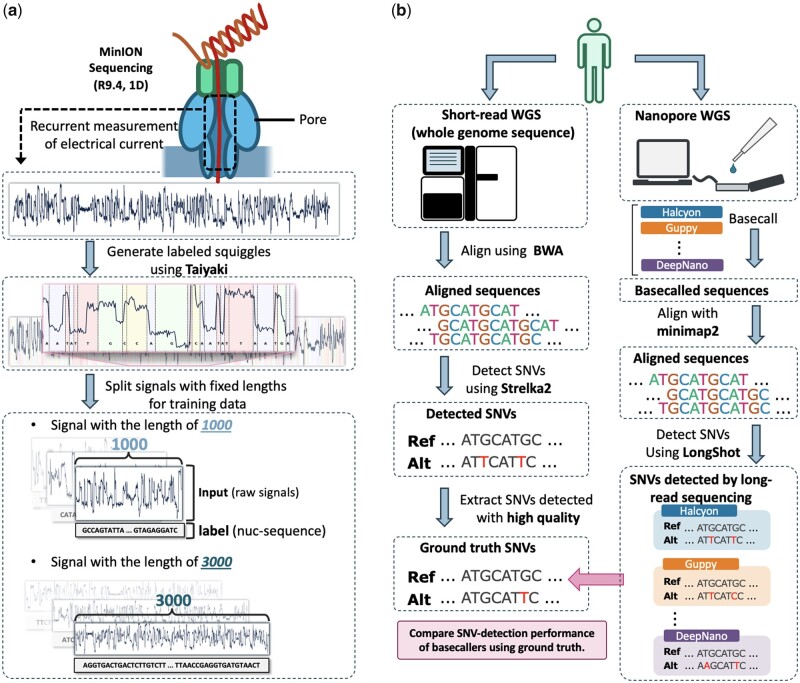
Fig. 2.
(a) Overview of preparation of training datasets using ONT’s retraining model, Taiyaki. Labeled reads obtained by Taiyaki are then split into fixed-length raw signals and corresponding nucleotide sequences. (b) Overview of evaluation of different basecallers in terms of SNV-detection performance assuming short-read sequencing as the ground truth