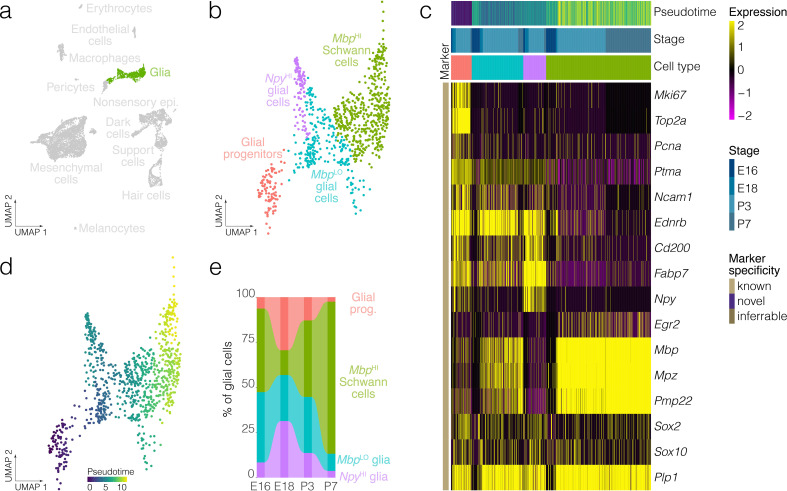
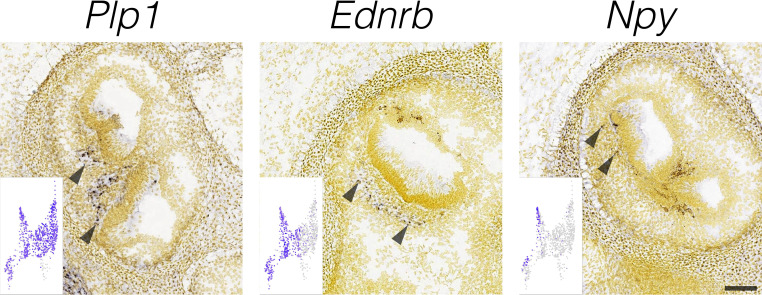
Figure 3. Glial cell diversity and dynamics.
(a) Highlights the position of glial cells in UMAP space relative to the whole dataset. (b) Cluster analysis in glia. (c) Expression of markers relative to cell cluster, developmental stage and pseudotime. (d) Pseudotime of the developmental trajectory in glia. (e) The relative contributions of cell clusters to the composition of glia at the indicated developmental stages. Note that Schwann cells and myelinating cell markers increase with the developmental time.