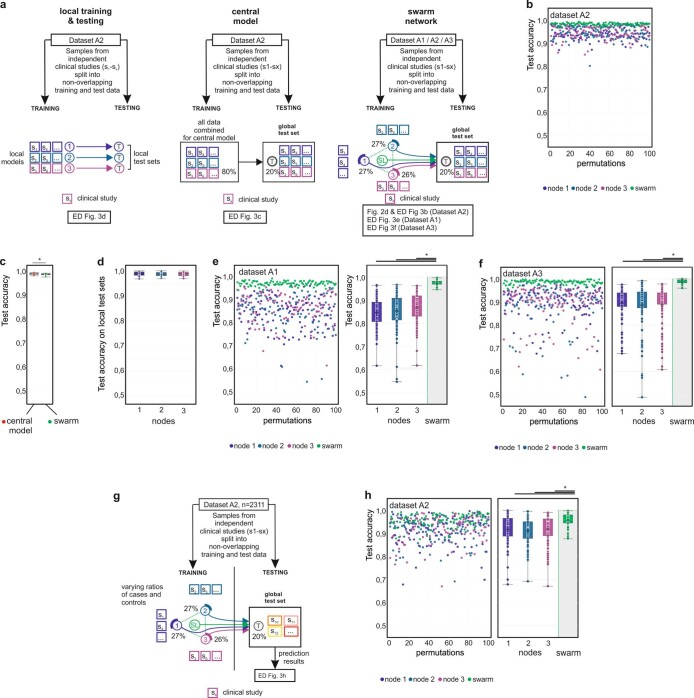
Extended Data Fig. 3. Scenario to test for batch effects of siloed studies in datasets A1–A3 and scenario with multiple consortia.
Main settings and representation of schema and data visualization are as in Fig. 2a. a, Scenario with training nodes coming from independent clinical studies for local models (left), central model (middle) and the Swarm network (right) and testing on a non-overlapping global test with samples from the same studies. b, Evaluation of test accuracy over 100 permutations for dataset A2 with the scenario shown in a (right) and Fig. 2d. c, Comparison of test accuracy between central model (a, middle) and SL (a, right). d, Comparison of test accuracy on the local test datasets (a, left) for 100 permutations. e, Evaluation of test accuracy of individual nodes versus SL over 100 permutations for dataset A1 when training nodes have data from independent clinical studies. f, Evaluation of test accuracy of individual nodes versus SL over 100 permutations for dataset A3 when training nodes have data from independent clinical studies. g, Scenario with three consortia contributing training nodes and a fourth one providing the testing node. h, Evaluation of test accuracy for scenario shown in g over 100 permutations for dataset A2. d–f, h, Box plots show representation of accuracy of all permutations performed for the 3 training nodes individually as well as the results obtained by SL (d only for local models). All samples are biological replicates. Centre dot, mean; box limits, 1st and 3rd quartiles; whiskers, minimum and maximum values. Performance measures are defined for the independent fourth node used for testing only. Statistical differences between results derived by SL and all individual nodes including all permutations performed were calculated with one-sided Wilcoxon signed rank test with continuity correction; *P < 0.05, exact P values are listed in Supplementary Table 5.