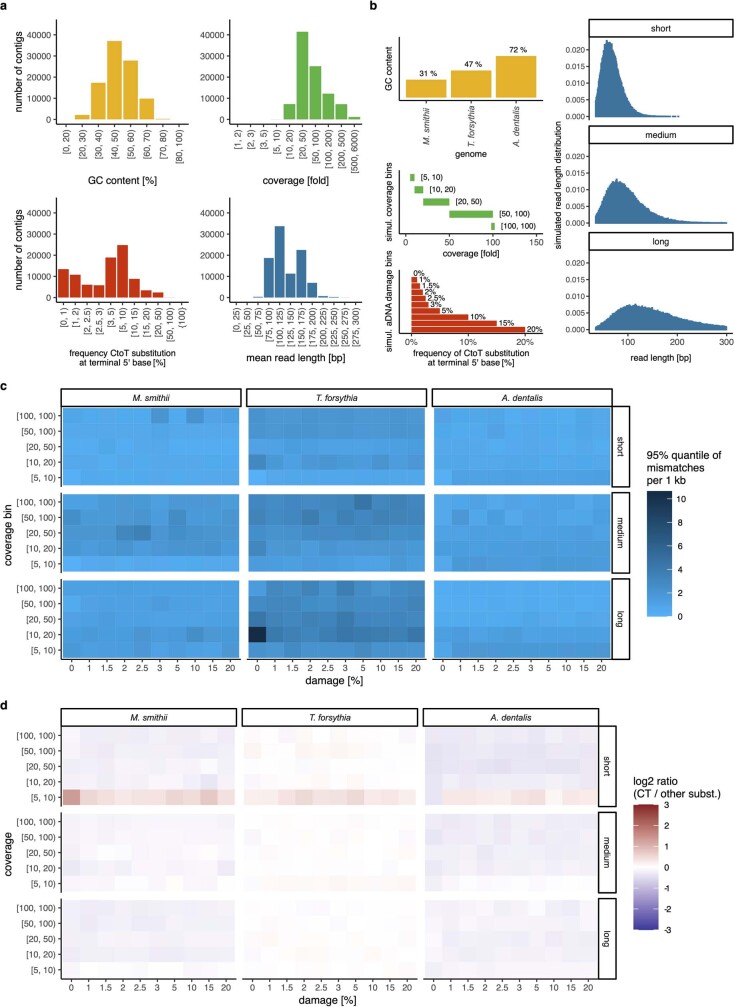
Extended Data Fig. 9. Effect of aDNA damage on the assembly of short-read data.
Related to Fig. 2, see Supplementary Information section 6. a, Distribution of the values of four sequencing data variables that may have an effect on the assembly of short-read data and were observed in the 498 medium-quality and high-quality MAGs assembled in this study. b, Overview of the parameter space of the variables GC content, sequencing depth, observed aDNA damage and read length that was used for simulating short-read sequencing using gargammel107. c, Number of mismatches per 1 kb of alignable contig sequence with respect to the reference genome as observed at the 95% quantile for all combinations of reference genome, read length distribution, simulated aDNA damage and coverage averaged across the five replicates. d, The log2-transformed ratio of C-to-T substitutions to the average number of all other substitutions per 1 kb of alignable contig sequence for all combinations of reference genome, read length distribution, simulated aDNA damage and coverage averaged across the five replicates. Positive values indicate an excess of C-to-T substitutions.