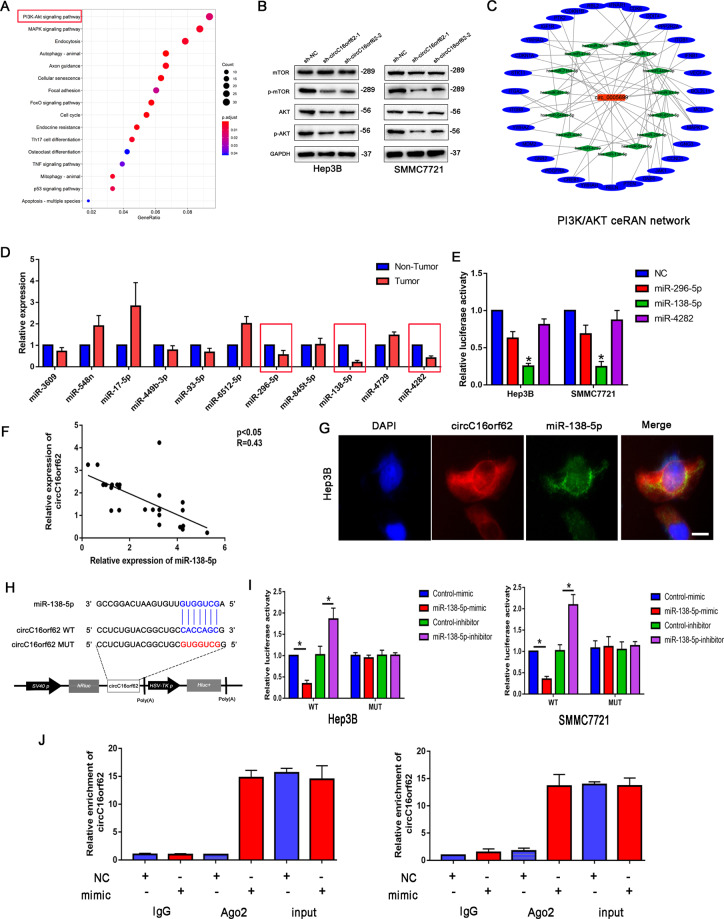
Fig. 4. Bioinformatic analysis and validation of potentially targeted miRNAs of circC16orf62 in HCC cells.
A KEGG pathway analysis the downstream signal pathway eluted the PI3K/AKT was the significantly downstream pathway of circC16orf62. B Western-blots analysis the expression of p-AKT and p-mTOR. C ceRNA network analysis the target miRNAs of circC16orf62. D RT-qPCR and western blot analysis the target miRNAs in the HCC tumor and adjacent normal tissue. E Luciferase analysis the interaction of circC16orf62 and miRNA-138. F RT-qPCR analysis the expression correlation of circC16orf62 and miRNA-138 in the HCC tumor and adjacent normal tissue. G FISH assay analysis the circC16orf62 and miRNA-138 co-location in the Hep3B cell, the scale bar represents 10 µm. H TargetScan showed that circC16orf62 could bind to the miR-138-5p. I The direct binding between circC16orf62 and miRNA-138 was analyzed by dual-luciferase reporter assay. J A RIP assay was performed using an AGO2 antibody, and IgG served as a negative control. Data are shown as the mean ± standard deviation of three independent experiments.*P < 0.05was regarded as statistically significant.