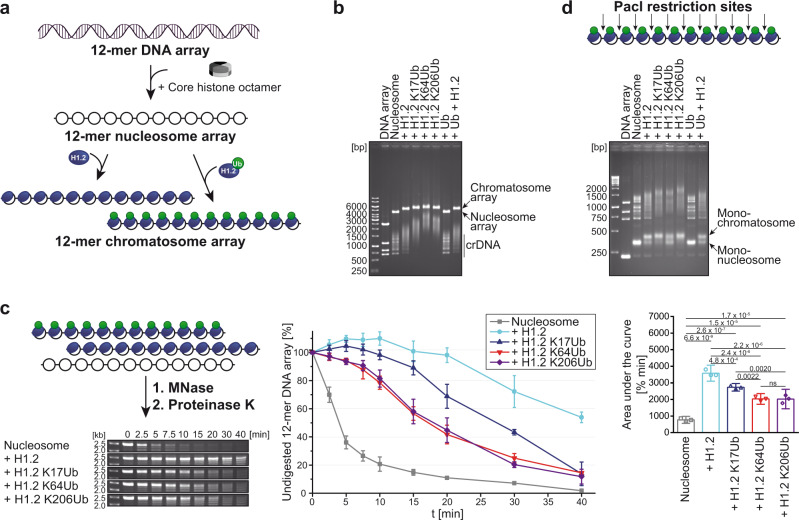
Fig. 4. Impact of H1.2 KxUb variants on chromatosome assembly.
a Schematic overview of 12-mer chromatosome array formation containing H1.2 or H1.2 KxUbs. b EMSA of different arrays analyzed; crDNA indicates competitor DNA. c Overview of MNase digestion assay which probes DNA accessibility of nucleosome and chromatosome arrays (left). The arrays were treated with MNase for indicated times, digested by proteinase K, and analyzed by agarose gel electrophoresis. Undigested arrays were quantified and plotted over time (middle). Shown are mean values +/− standard deviation, n = 3. For statistical analysis, the area under the curve for the digestion of the different arrays was determined and analyzed (right); shown are mean values +/− standard deviation, n = 3, one-way ANOVA with Tukey’s multiple comparisons test with α = 0.05, 95% confidence interval, exact p values with p ≤ 0.05 are shown, ns (not significant) p > 0.05. d PacI digestion assay. PacI cleaves the array site-specifically within the linker DNA between the repetitive nucleosome positioning sequences resulting in the formation of mono-nucleosomes and mono-chromatosomes.