Figure 5.
Gene expression profiling reveals a common transcriptional program between freshly isolated and hibernating HSCs
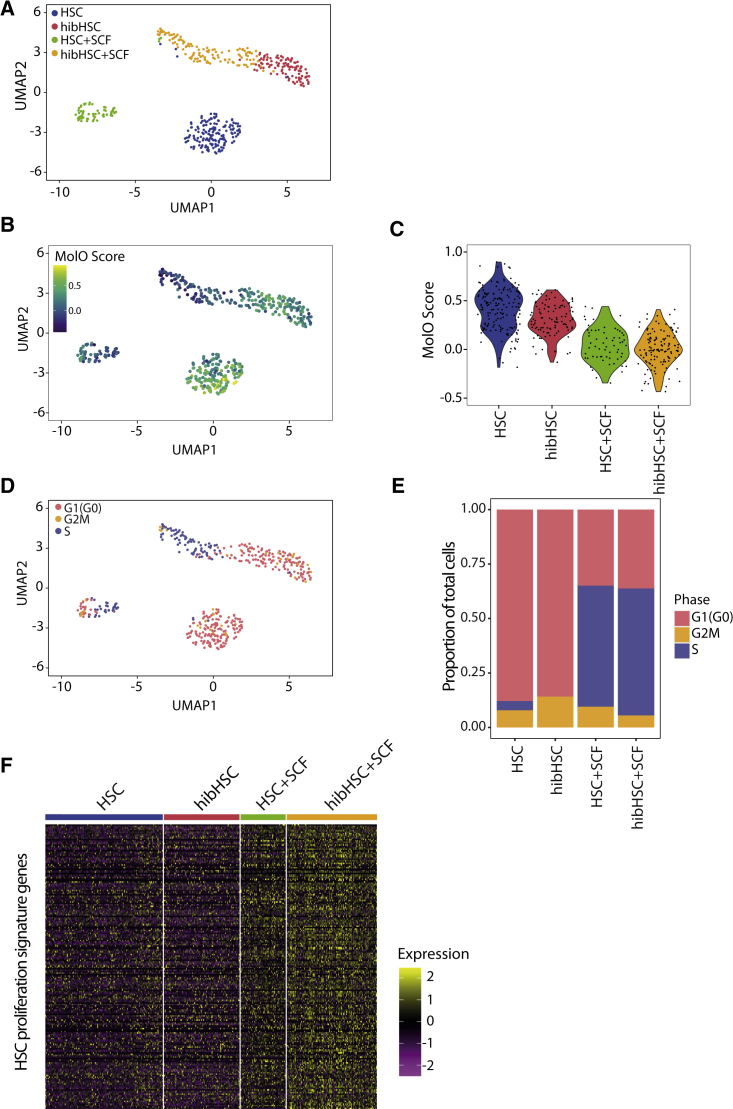
(A) Uniform Manifold Approximation and Projection (UMAP) of scRNA-seq profiles derived from four distinct populations (HSC, blue dots; hibHSC, red dots; HSC + SCF, green dots; hibHSC + SCF, orange dots).
(B) The HSC-specific Molecular Overlap (MolO) gene signature score was computed based on average expression of signature genes and projected onto the UMAP distribution.
(C) MolO scores for the individual HSCs in each physiological state with the HSCs and hibHSCs having the highest overall scores.
(D) Cell-cycle scores were computed for each cell and identified states were projected on the UMAP display from 5A (G1(G0), pink; G2/M, orange; S, blue).
(E) A proportional representation of cell-cycle stages of all cells within each distinct population (G1(G0), pink; G2/M, orange; S, blue).
(F) Heatmap of previously identified HSC-specific proliferation signature genes (Venezia et al., 2004) sorted by cell type with low expression in HSCs and hibHSCs and high expression in both sets of SCF-stimulated cells.