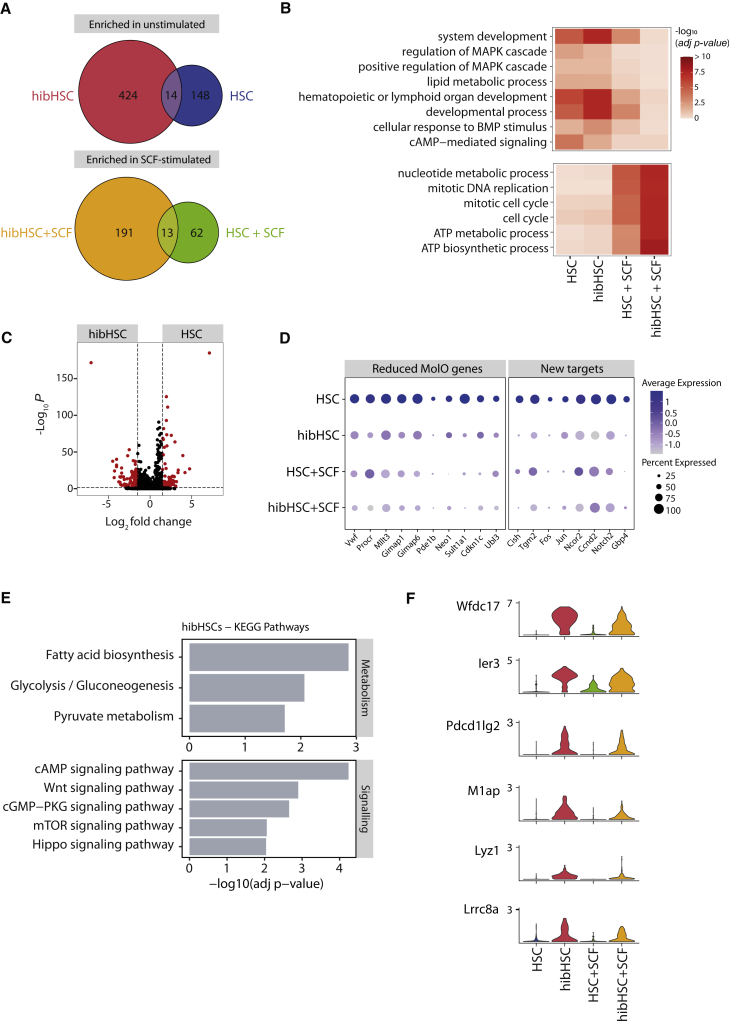
Figure 6.
Hibernating HSCs have a unique molecular profile of stress response
(A) Differential gene expression (DGE) was computed for two separate comparisons: (1) comparison of fresh HSCs (HSC) against SCF-stimulated HSCs (HSC + SCF); (2) comparison of hibernating HSC (hibHSCs) against hibHSCs post-SCF-stimulation (hibHSC + SCF) (negative binomial distribution, adjusted with Benjamini-Hochberg correction). Venn diagrams represent the number of genes commonly enriched in unstimulated populations (HSC and hibHSC) and SCF-stimulated populations (HSC + SCF and hibHSC + SCF) from both separate DGE computations.
(B) Gene ontology term enrichment was computed based on differentially expressed genes, as outlined in (A). Minimum p value > 0.05 to be considered significantly enriched.
(C) Volcano plot of differentially expressed genes (red dots), comparing fresh HSCs (HSC) and hibernating HSCs (hibHSC) (negative binomial distribution, adjusted with Benjamini-Hochberg correction).
(D) Dot plot representing the average normalized expression of genes across the four distinct populations. Genes of interest and MolO signature genes were selected from DGE in (C). The size of each dot indicates the proportion of cells with normalized expression level >0 (scaled expression represented by color intensity).
(E) KEGG pathway enrichment in unstimulated hibernating HSCs (hibHSC), showing selected metabolic and signal transduction pathways (enrichment cutoff: adjusted p value > 0.05).
(F) Violin plots of normalized gene expression of selected differentially expressed genes, enriched in unstimulated hibernating HSCs (hibHSCs).