Figure 7.
In silico analysis of miRNA/mRNA interactions
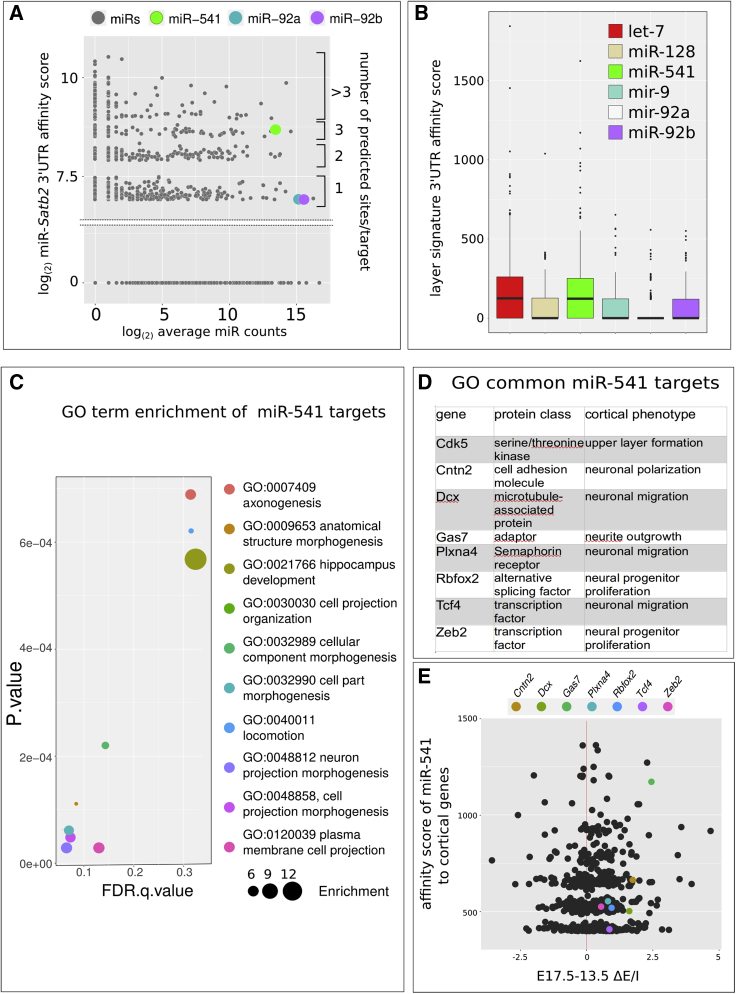
(A) In silico comparison of the affinity of mouse miRNAome (gray dots), miR-92a/b, and miR-541 (colored dots) with Satb2 3′UTR (Ensembl Mus musculus Satb2-201 cDNA 3′UTR), in relation to the average miRNA expression levels during corticogenesis.
(B) In silico affinity of cortical miRNAs to the 3′UTR of an embryonic cortical layer gene signature (395 genes) (Galfrè et al., 2020).
(C) GO enrichment of the mir-541 gene targets with high in silico affinity to Satb2 3′UTR (cumulative score higher than 400, n = 48) (Enright et al., 2003) with respect to the layer gene signature employed in (B).
(D) List of the eight genes common to all the GO terms shown in (C).
(E) Plot showing E/I read counts developmental increase (x axis) with respect to miR-541/mRNA affinity score (y axis) to genes of the embryonic cortical marker signature (Galfrè et al., 2020). Colored dots indicate genes listed in (D). Names in labels indicate the five genes with the highest E/I read count ratio increase and mir-541/mRNA affinity score.