Fig. 1. Reconstitution of valinomycin biosynthetic pathway in vitro.
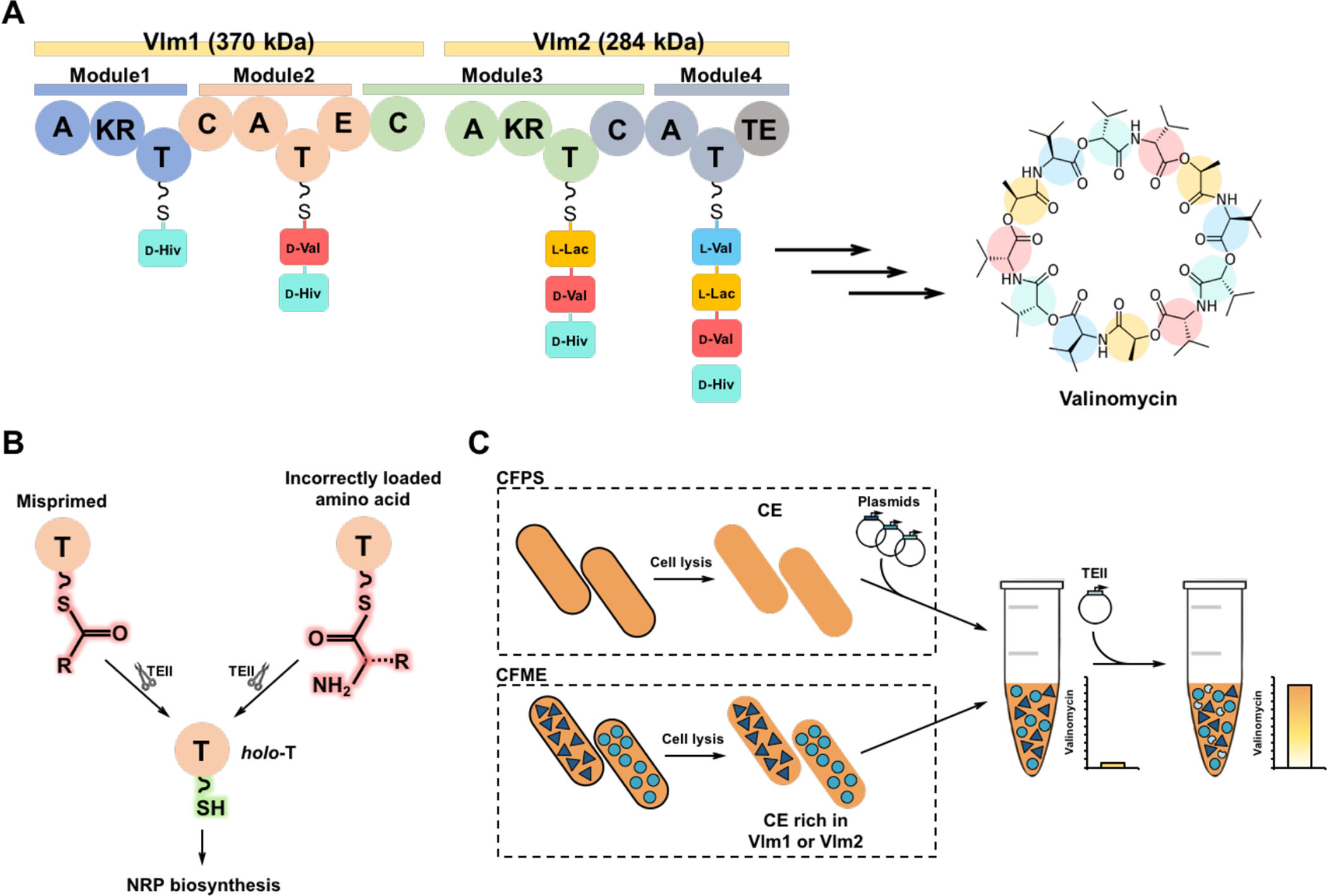
(A) Valinomycin synthetase contains two distinct NRPS enzymes Vlm1 (374 kDa) and Vlm2 (284 kDa) that are composed of adenylation (A), ketoreductase (KR), thiolation (T), condensation (C), epimerase (E), and thioesterase (TE) domains. A domains select and activate three substrates (pyruvate, α-ketoisovalerate (α-Kiv), and l-valine (l-Val)). T domains are responsible for the translocation of the bound aminoacyl or peptidyl intermediate between adjacent catalytic positions. C domains catalyze the formation of peptide bond and elongate the peptide chain. The KR domain in Module 1 reduces α-Kiv to d-hydroxyisovalerate (d-Hiv). The E domain in Module 2 converts l-Val to d-valine (d-Val). The KR domain in Module 3 reduces pyruvate to l-lactate (l-Lac). The four modules of valinomycin synthetase are iteratively reused to assemble three tetradepsipeptide monomers, which are eventually oligomerized and macrolactonized to form the 36-membered cyclododecadepsipeptide valinomycin. (B) Regeneration of the functionality of T domains in NRPS catalyzed by type II thioesterase (TEII). (C) Cell-free biosynthesis of valinomycin with different strategies: cell-free protein synthesis (CFPS), cell-free metabolic engineering (CFME), and a coupled CFPS-MS approach. CE, cell extract.
