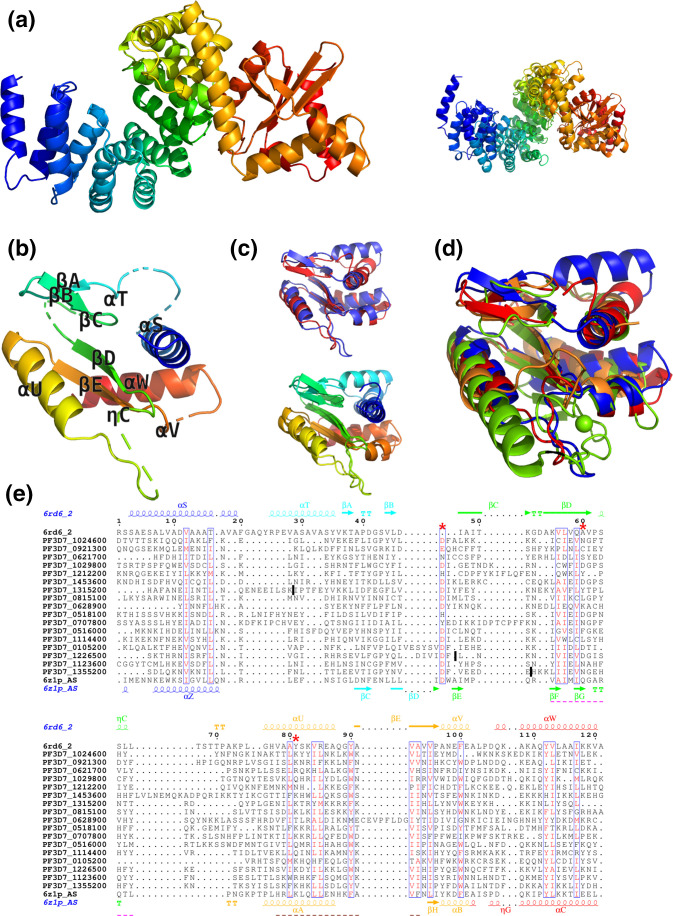
Fig. 5.
Structure predictions for RAP proteins from Plasmodium falciparum. (a) Left: the modelling template covering both HPRs and RAP domain from Plasmodium falciparum proteins, based on experimental structure of F-ATP synthase from Polytomella sp. Pringsheim 198.80 (PDB code: 6rd6 chain 2, residues 8–445). Right: the superposition of the two available modelling templates covering these domains (6rd6 chain 2 and 6z1p chain AS). (b) Structural model of the RAP domain of PF3D7_1024600 (residues 327–431), based on 6rd6 chain 2. (c) Top: the structural superposition of the two closest modelling templates for the RAP domain from PF3D7_1024600 6rd6 chain 2 residues 322–445 (blue) and 6z1p chain AS residues 591–685 (red). Bottom: the same superposition is shown in rainbow N-to-C terminus colouring. (d) Structural superposition of different modelling templates used for the RAP domain: 6rd6 chain 2 residues 322–445 (blue), 6z1p chain AS residues 591–685 (red), 3r3p chain A (orange) and 1vsr chain A (green). (e) Alignment of RAP domains from Plasmodium falciparum and F-ATP synthase (6rd6_2). The sequences and secondary structure from the two closest modelling templates are shown in the top and bottom rows of the alignment. The most conserved parts of the first and the second sequence motifs identified in Apicomplexan RAP domains (see Fig. 4, top row) are underlined with pink and magenta dashed lines, respectively. The red asterisks indicate the three residues of the PD-(D/E)XK superfamily. Positions of removed repeats are marked by dark vertical bars.