Fig. 1.
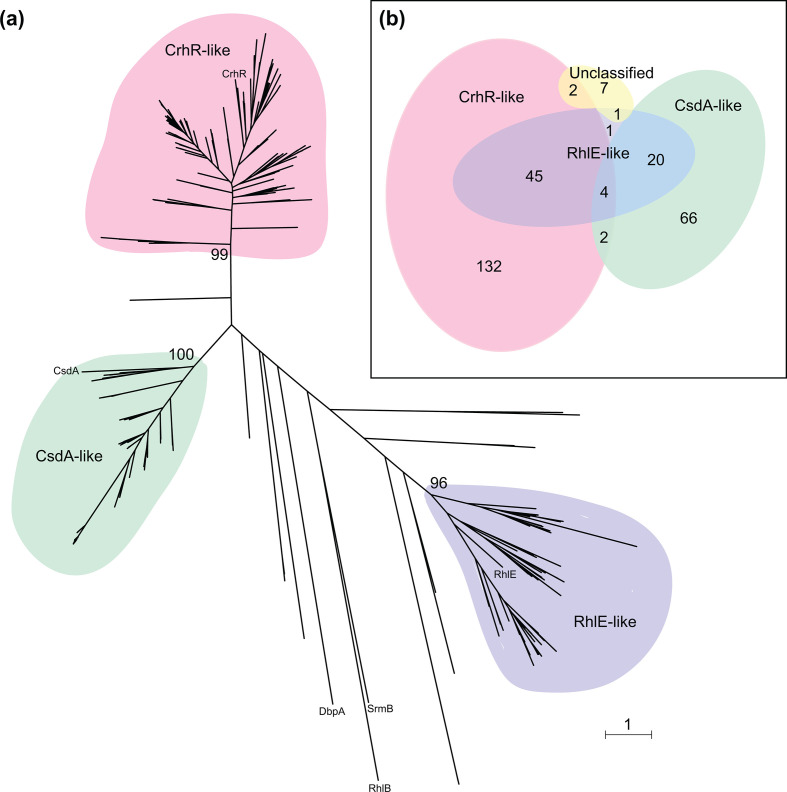
Cyanobacterial DEAD-box proteins cluster into three clades. The phylogenetic tree (a) shows the 362 cyanobacterial DEAD-box proteins form three major clades with bootstrap values ≥95 %, corresponding to CsdA-like and RhlE-like proteins, and a clade unique to cyanobacteria, CrhR-like proteins, named for the sole DEAD-box RNA helicase encoded in the genome of Synechocystis sp. PCC 6803. Bootstrap values at the root of each clade, as well as the positions of the five DEAD-box proteins of E. coli and CrhR from Synechocystis sp. PCC 6803, are indicated. Branch lengths are proportional to the mean number of substitutions per amino acid site as indicated by the scale bar. The distribution of the DEAD-box protein clades identified in the 280 cyanobacterial strains used in the analysis is represented by the Euler diagram shown in the inset (b).