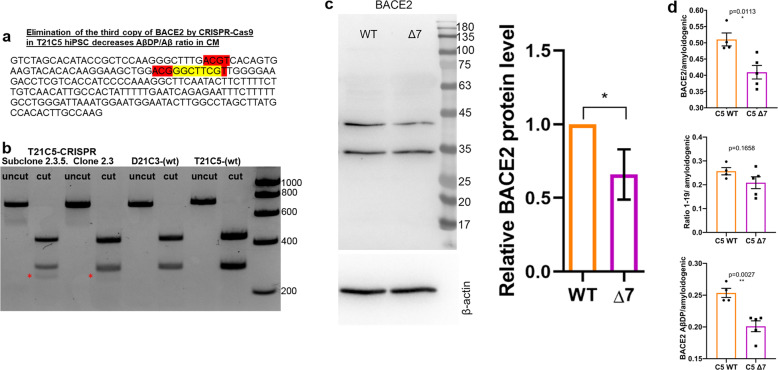
Fig. 5. CRISPR/SpCas9-HF1-mediated reduction of BACE2 copy number from three to two in the T21C5 hiPSC line.
a BACE2 exon3 sequence with 7 bp deletion (yellow) provoked by the CRISPR/SpCas9-HF1 is shown. Red: restriction endonuclease HpyCH4IV sites (a de novo HpyCH4IV site is generated by the 7 bp deletion). b agarose gel electrophoresis of the 733 bp PCR product containing the targeted site before (uncut) and after digestion with HpyCH4IV (cut), for the initial clone 2.5, and its colony-purified sub-clone 2.3.5 (renamed further below as “Δ7”). The 294 bp fragment in 2.3.5 is reduced to 65% of the wt value (normalized to the 439 bp band), and a de novo 255 bp fragment appears in CRISPR-targeted line (red asterisk). c Western blot stained with anti-BACE2 antibody of the lysates of the iPSC line Δ7 compared with the wt T21C5 iPSC line. Quantification of the total actin-normalised BACE2 signal showed a significant reduction in Δ7 compared with T21 unedited line. Error bars: standard error, p value: student’s t test. d BACE2-AβDP/amyloidogenic peptides ratio after IP-MS analysis of CM produced by the 48DIV organoids derived from the iPSC line Δ7 compared with the T21C5wt control were significantly decreased. Error bars: standard error, p values: two-tailed t test comparison.