Abstract
Increasing evidence associates indoor fungal exposure with deleterious central nervous system (CNS) health, such as cognitive and emotional deficits in children and adults, but the specific mechanisms by which it might impact the brain are poorly understood. Mice were exposed to filtered air, heat-inactivated Aspergillus versicolor (3 × 105 spores), or viable A. versicolor (3 × 105 spores) via nose-only inhalation exposure 2 times per week for 1, 2, or 4 weeks. Analysis of cortex, midbrain, olfactory bulb, and cerebellum tissue from mice exposed to viable A. versicolor spores for 1, 2, and 4 weeks revealed significantly elevated pro-inflammatory (Tnf and Il1b) and glial activity (Gdnf and Cxc3r1) gene expression in several brain regions when compared to filtered air control, with the most consistent and pronounced neuroimmune response 48H following the 4-week exposure in the midbrain and frontal lobe. Bulk RNA-seq analysis of the midbrain tissue confirmed that 4 weeks of A. versicolor exposure resulted in significant transcriptional enrichment of several biological pathways compared to the filtered air control, including neuroinflammation, glial cell activation, and regulation of postsynaptic organization. Upregulation of Drd1, Penk, and Pdyn mRNA expression was confirmed in the 4-week A. versicolor exposed midbrain tissue, highlighting that gene expression important for neurotransmission was affected by repeated A. versicolor inhalation exposure. Taken together, these findings indicate that the brain can detect and respond to A. versicolor inhalation exposure with changes in neuroimmune and neurotransmission gene expression, providing much needed insight into how inhaled fungal exposures can affect CNS responses and regulate neuroimmune homeostasis.
Keywords: Aspergillus versicolor, filamentous fungi, microglia, neuroinflammation, neuroimmune homeostasis. neuropeptides, RNA-seq
Introduction
Fungal bioaerosols in indoor environments are a common and deleterious exposure that is associated with several respiratory diseases such as asthma and allergic rhinitis (Heseltine et al., 2009). Further, several small studies suggest that individuals who spend time in fungal contaminated environments also report anxiety, cognitive dysfunction, and “brain fog” (Lieberman et al., 2006; Ratnaseelan et al., 2018; Hyvonen et al., 2020). Other small studies have suggested that patients who have been repeatedly exposed to fungi have cognitive and emotional impairment similar to patients with mild to moderate traumatic brain injury (Baldo et al., 2002; Gordon et al., 2004). However, there is significant controversy surrounding the study design and small sample sizes of these reports (McCaffrey & Yantz, 2005). Given that the average individual in the U.S. spends an estimated 87% of their time indoors (Klepeis et al., 2001), it is critical that indoor pollutants such as fungal exposure be investigated for their extra-pulmonary, and specifically central nervous system (CNS) effects. Currently, the mechanisms underlying how fungal exposure may impact the CNS are poorly understood.
Certain filamentous fungi can thrive in damp environments, especially water-damaged homes and buildings (Heseltine et al., 2009). While the pulmonary consequences of fungal exposure have been extensively studied in both human and animal studies (Etzel & Rylander, 1999; Mendell et al., 2011; Vesper & Wymer, 2016; Nayak et al., 2018; Croston et al., 2020), few studies have examined the impact of filamentous fungi on the CNS. Interestingly, a recent report investigating the CNS consequences of exposure to a fungal particulate mixture revealed that exposed mice had an attenuated neuroimmune transcriptional response in brain stem regions controlling respiration (Peng et al., 2018). An additional study has defined how intranasal exposure to live and heat inactivated Stachybotrys chartarum spores, a potent toxin-producing species commonly known as “black mold”, can augment expression of pro-inflammatory markers in the brain and induce behavioral deficits in mice (Harding et al., 2020). However, no studies have investigated how other, more common and ubiquitous fungal species affect the CNS. In particular, Aspergillus spp. are among the most common indoor fungi and are associated with the musty odor of water-damaged buildings (Schweinsberg & Mersch-Sundermann, 2003; Heseltine et al., 2009). Interestingly, Aspergillus versicolor, found in greater than 75% of water-damaged buildings (Beguin & Nolard, 1994), has been shown to have robust and differential immune effects in both human cells and rodents, where A. versicolor induces a Th17 polarization, and A. versicolor extracts can be directly cytotoxic to immune cells (Mintz-Cole et al., 2012; Pei & Gunsch, 2013). While evidence points to potential CNS effects in response to fungal exposure, the CNS impact of viable filamentous fungi, the neuroimmune consequences for A. versicolor, and the neurochemical sequelae that could be modifying behavior are largely unknown.
At present, it has been well established that outdoor air pollution exposure, an inhaled toxicant, has neuroimmune consequences (Calderón-Garcidueñas et al., 2004; Campbell et al., 2005; Calderón-Garcidueñas et al., 2008; Campbell et al., 2009; Calderón-Garcidueñas et al., 2012; Tyler et al., 2016; Costa et al., 2017; Klocke et al., 2017). Often associated with CNS disease and damage, neuroinflammation can be broadly defined as the elevation of cytokines and reactive oxygen species in the CNS (Costa et al., 2017). Neuroinflammation often occurs when the brain detects inhaled toxins/toxicants, where neuroinflammation is proposed as a common mechanism through which inhaled environmental exposures may affect CNS function and health (Costa et al., 2017; Jayaraj et al., 2017). In the current study, we will begin to assess how A. versicolor inhalation exposure affects the brain by characterizing the CNS transcriptional signature and exploring the temporal requirements necessary to trigger neuroinflammation.
Methods
Reagents
The polyclonal rabbit anti-ionized calcium binding adaptor molecule 1 (IBA-1) antibody (Wako Cat# 019-19741, RRID:AB_839504 Lot#: CAK1997) was purchased from Wako (Richmond, VA). The rabbit anti-doublecortin (DCX) polyclonal antibody (Cat# 4604, RRID:AB_561007) was acquired from Cell Signaling Technologies (Danvers, MA). The biotinylated goat anti-rabbit secondary antibody (Vector Laboratories Cat# BA-1000, RRID:AB_2313606 Lot#: ZB1007) was purchased from Vector Laboratories (Burlingame, CA). The Alexa Fluor 488 goat anti-rabbit IgG (Thermo Fisher Scientific Cat# A31566, RRID:AB10374301) was purchased from Thermo Fisher Scientific (Waltham, MA). All other reagents were procured from Sigma-Aldrich (St. Louis, MO).
Animals
Five to six week old female B6C3F1/N mice, a standard immunotoxicity model for filamentous fungi exposure (King-Herbert & Thayer, 2006; Nayak et al., 2018), were acquired from NTP/Taconic (Germantown, New York) and were acclimated for two weeks prior to inhalation exposures. Groups of five mice were housed in HEPA-filtered, ventilated polycarbonate cages (Lab Products, Inc., Seaford, DE) and were provided an NTP-2000 diet (Harlan Laboratories) and tap water ad libitum. The National Institute for Occupational Safety and Health (NIOSH) animal facility is an environmentally-controlled barrier facility that is fully accredited by the AAALAC International. All animal procedures were performed under the CDC-Morgantown Institutional Animal Care and Use Committee approved protocol18-008.
A total of 126 mice were assigned to treatment groups in a randomized block design (Table 1), with 7 mice per group. Mice underwent the predetermined inhalation exposure for the specified length and brain tissue was collected for analysis at either 24 H or 48 H after the last exposure. No animals were excluded nor died during the experiment. Samples from one experimental animal was sufficient for RNA, protein, and the immunohistochemical endpoints measured.
Table 1.
Experimental Design.
| Exposure length | Time post exposure |
Inhalation exposure |
||
|---|---|---|---|---|
| FA | HIC | AV | ||
| 1 Week | 24 h | n = 7 | n = 7 | n = 7 |
| 48 h | n = 7 | n = 7 | n = 7 | |
| 2 Week | 24 h | n = 7 | n = 7 | n = 7 |
| 48 h | n = 7 | n = 7 | n = 7 | |
| 4 Week | 24 h | n = 7 | n = 7 | n = 7 |
| 48 h | n = 7 | n = 7 | n = 7 | |
A total of 126 mice were assigned to treatment groups in a randomized block design, with 7 mice per group. Mice underwent the predetermined inhalation exposure for the specified length and brain tissue was collected for analysis at either 24 H or 48 H after the last exposure. No animals were excluded and samples from one experimental animal was sufficient for RNA, protein, and the immunohistochemical endpoints measured. Filtered Air (FA);Heat Inactivated Conidia(HIC);Viable Aspergillus versicolor (AV).
Aspergillus versicolor Inhalation Exposure
The primary route of human filamentous fungi exposure occurs when airborne fungal bioaerosols, comprised primarily of spores and hyphal fragments, are inhaled. Many early fungal exposure models in mice employed the delivery of poorly characterized crude fungal extracts or liquid conidia suspensions using involuntary aspiration and intranasal approaches. These methods do not consistently reproduce the pulmonary pathology associated with environmentally relevant chronic exposures (Nayak et al., 2016). To accurately model A. versicolor exposure, mice were exposed to A. versicolor using a nose-only, acoustical generator exposure system (AGS) that allows for real-time analysis of particle size, deposition estimations, and manipulation of exposure concentrations (Buskirk et al., 2014). Specifically, the acoustical generator system delivers dry aerosolized fungal test articles to mice housed in nose-only chambers (Buskirk et al., 2014). The acoustical generator system employs an algorithm with a scale factor accounting for the conidial frequency within the different aerodynamic particle size bins, the DataRAM’s mass concentration measurements, and a mouse minute volume (25 mL) to estimate the real-time number of conidia deposited within the murine lung (Buskirk et al., 2014). Once the targeted concentration of the estimated deposited conidia is achieved, the system switches to HEPA-filtered air until the exposure time of 1 H has lapsed, as previously published (Buskirk et al., 2014). The time of exposure of 1 H was previously determined to reduce variability in the sporulation and aerosolization of the test article, allowing reliable and reproducible targeted concentrations of deposited conidia in the lung (Buskirk et al., 2014).
To generate the exposure particles, A. versicolor spores (strain ATCC 9577, (Vuillemin) Tiraboschi, American Type Culture Collection, Manassas, VA, USA) were first cultured on a malt extract agar and harvested after 7–8 days. Spores were then resuspended in filter sterilized distilled water (HyClone Laboratories Inc., South Logan, Utah), and cultured on autoclaved rice for 7–10 days. Heat-inactivated (HIC) A. versicolor serve as a particle control and were generated by baking cultures at 80°C for 4 hours.
Mice were acclimatized to the AGS for one week and then were exposed to either HEPA filtered air (FA), 3 × 105 spores of HIC A. versicolor, or 3 × 105 spores of viable A. versicolor (AV) twice a week for 1, 2, or 4 weeks, using the AGS as previously described (Buskirk et al., 2014). The spore exposure level is reported as the estimated pulmonary spore deposition per exposure. Animals were euthanized and brain tissue was obtained 24H or 48H after the last A. versicolor exposure. The welfare of experimental mice was closely monitored with daily health checks and weights were recorded once per week for the duration of the exposure study. No adverse effects were identified during the study. Following the final A. versicolor exposure, animals were humanely euthanized with 100–300 mg/kg sodium pentobarbital euthanasia solution and exsanguinated via cardiac puncture. Pulmonary inflammation and spore deposition were documented with histology for the 4 week exposure (Figure 1). Brain tissue was then obtained 24H or 48H after the final exposure, as it has been previously determined that pulmonary inflammation in response to filamentous fungi using this exposure system persists to 48H after the exposure (Buskirk et al., 2014). The right hemisphere was fixed in 4% paraformaldehyde and the left hemisphere tissue was snap frozen and stored at −80°C. The olfactory bulb, frontal lobe, temporal lobe, cerebellum, and midbrain were dissected from frozen tissue of the left hemisphere.
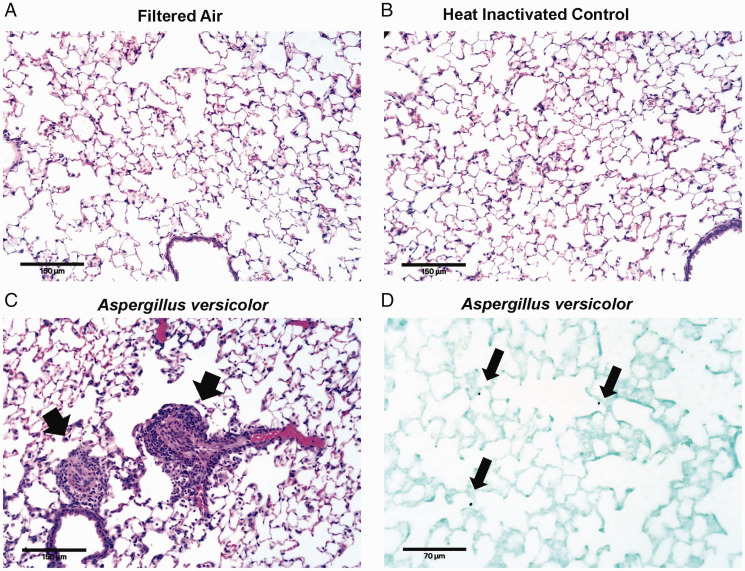
Figure 1.
Aspergillus versicolor exposure causes pulmonary inflammation. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks. Representative photomicrographs of hematoxylin eosin staining of murine lung sections are shown (A–C). Airway inflammation (indicated by the black arrowheads) was observed following exposure to viable A. versicolor and not FA nor HIC. Images were captured using a 20× objective and the scale bar indicates 100 µM. A representative image of Grocott's methenamine silver staining in lung tissue from mice exposed to live AV (D). Black arrows indicate conidia deposited in the lung. Images were captured using a 40× objective and the scale bar indicates 70 µM.
Lung Histology Methods
To confirm spore deposition and pulmonary inflammation, murine lungs were instilled with 10% neutral-buffered formalin, paraffin embedded and sectioned at 5 microns. Sections were stained with hematoxylin and eosin for histopathology evaluation pulmonary inflammation. Representative photomicrographs were captured using an Echo Rebel Hybrid Microscope (Echo, San Diego, CA) using a 20× plan achromat objective. For evaluation of fungal spore deposition, mice were exposed to a single-dose of viable A. versicolor (1 × 105) and sacrificed immediately following exposure. Murine lungs were processed as above and stained with Grocott's methenamine silver stain. Representative photomicrographs were captured using an Echo Rebel Hybrid Microscope using a 40× plan achromat objective.
Immunohistochemistry – Chromogenic Detection
The right hemisphere of the brain was fixed in 4% paraformaldehyde for 2 days and cryoprotected in 30% sucrose. Coronal sections (40 µm) were collected using a freezing stage microtome (Microm HM 450, Thermo Scientific, Waltham, MA). Free-floating sections were treated with 1% hydrogen peroxide, washed three times for 10 minutes with phosphate buffered saline (PBS), incubated 1Hin blocking solution (PBS containing 1% bovine serum albumin, 4% goat serum, 0.4% Triton X-100) and incubated overnight at 4°C with primary antibodies diluted in DAKO antibody diluent (Agilent, Santa Clara, CA). IBA-1 and DCX antibodies were used at a 1:1,000 dilution to stain microglia and immature neurons, respectively. Sections were then washed three times in PBS, incubated with biotinylated anti-rabbit antibody for 1H, washed three times in PBS, and incubated with Vectastain ABC Kit (Vector Laboratories, Burlingame, CA) reagents according to manufacturer’s instructions. Staining was visualized using 3,3′-diaminobenzidine (DAB) and urea-hydrogen peroxide tablets (Sigma-Aldrich, St. Louis, MO). Images were captured with a Leica DM2500 microscope (Leica Microsystems, Buffalo Grove, IL). Slides were blinded and cells were counted by a blinded observer.
Unbiased Stereology – Microglia and Doublecortin (DCX) Positive Neuron Cell Number
Microglia and DCX+ neuron counts were acquired using the optical fractionator method of unbiased stereology, with the Leica DM 2500 microscope, as previously described (Hutson et al., 2011). DCX+ neurons and IBA-1+ microglia were counted by a blind observer in the dentate gyrus of the hippocampus using 8 slices of every 3rd interval, centered on bregma −2.70. Representative images were acquired at 40×.
Fluorescent Microglia Morphology Quantification
To assess changes in microglial cell volume, four coronal sections per mouse were stained with IBA-1 (1:1,000), and Z-stacks (1 µm steps, 40× objective) were acquired the sections using a Nikon A1R Confocal microscope (Nikon Instruments, Melville, NY) in the cortex, midbrain, and CA1 region of the hippocampus. Images were analyzed using NIS Elements AR (Nikon Instruments) using the general analysis 3 module. Three-dimensional (3D) images were first thresholded to determine IBA-1 positivity and segmentation of individual microglia, and then the same threshold settings were propagated to all images in the experiment. After thresholding, 3D images were analyzed using the 3D processing function in NIS Elements AR to determine the volume of each individual microglia. Hypertrophic microglia were then defined as microglia with a volume of greater than 500 µm3. The number of hypertrophic microglia per stack were averaged, and then the stack average per slice were averaged to determine the microglial volume per animal.
RNA Extraction and Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the olfactory bulb, the frontal lobe, the cerebellum, and the midbrain using TRIzol (Invitrogen Life Technologies, Grand Island, NY), according to the manufacturer’s instructions. RNA was treated with Ambion DNA-free kit (Invitrogen Life Technologies, Grand Island, NY) according to manufacturer’s instructions. RNA was reverse transcribed using Maxima Reverse Transcriptase (Thermofisher Scientific, Waltham, MA). qRT-PCR was then performed using 1 µl of cDNA with Power Up SYBR Green Master Mix (Thermofisher Scientific, Waltham, MA), and 500 nM forward and reverse primers on a ViiA 7 qRT-PCR detection system (Life Technologies, Carlsbad, CA). Cycling parameters for all primers were set at 95°C for 5 minutes, followed by 40 cycles of 95°C (5 seconds)and 56°C (20 seconds), followed by a melt curve consisting of 5 second 0.5°C incremental increases from 65°C to 95°C. Cycles above 35 were considered undetectable. Values were normalized to Gapdh using the 2-ΔΔCT method. Primer sequences are listed in Table 2.
Table 2.
Syber Green Primer Sequences.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Gapdh | 5′-CCAGTGAGCTTCCCGTTCA-3′ | 5′-GAACATCATCCCTGCATCCA-3′ |
| Tnf | 5′-GCCCACGTCGTAGCAAACCACC-3′ | 5′-CCCATCGGCTGGCACCACTA-3′ |
| Cx3cr1 | 5′-TTCCCATCTGCTCAGGACCTC-3′ | 5′-GGTTCCAAAGGCCACAATGTC-3′ |
| Il1b | 5′-TGAAGAAGAGCCCATCCTCTGTGA-3′ | 5′-GGTCCGACAGCACGAGGCTT-3′ |
| Gdnf | 5′-GGATGGGATTCGGGCCACT-3′ | 5′-AGCCACGACATCCCATAACTTC-3′ |
Gapdh = Glyceraldehyde 3-phosphate dehydrogenase; Tnf = Tumor necrosis factor α; Cx3cr1 = CX3C chemokine receptor 1 (fractalkine receptor); Il1b = Interleukin 1β; Gdnf = Glial cell derived neurotrophic factor.
Midbrain mRNA Sequencing
Total RNA extracted from the midbrain was first evaluated for its quantity, and quality, using Agilent Bioanalyzer 2100. All RNA samples possessed an RNA integrity number (RIN) of 7 or higher. One hundred nanograms of total RNA were used. cDNA library preparation included mRNA purification/enrichment, RNA fragmentation, cDNA synthesis, ligation of index adaptors, and amplification, following the KAPA mRNA Hyper Prep Kit Technical Data Sheet, KR1352 – v4.17 (Roche Corporate). Each resulting indexed library was quantified and its quality accessed by Qubit and Agilent Bioanalyzer, and multiple libraries pooled in equal molarity. The pooled libraries were then denatured, and neutralized, before loading to NovaSeq 6000 sequencer at 300pM final concentration for 100 b paired-end sequencing (Illumina, Inc.). Approximately 30–40 M reads per library were generated. A Phred quality score (Q score) was used to measure the quality of sequencing. More than 95% of the sequencing reads reached Q30 (99.9% base call accuracy).
Mapping QC and Data Analysis
The sequencing data were first assessed using FastQC (Babraham Bioinformatics, Cambridge, UK) for quality control. Then all sequenced libraries were mapped to the mouse genome (mm10) using STAR RNA-seq aligner (v.2.5) (Dobin et al., 2013) with the following parameter: “–outSAMmapqUnique 60”. The reads distribution across the genome was assessed using bamutils (from ngsutils v.0.5.9) (Breese & Liu, 2013). Uniquely mapped sequencing reads were assigned to mm10 refGene genes using featureCounts (from subread v.1.5.1) (Liao et al., 2014) with the following parameters: “-s 2 –p –Q 10”. Differential expression analysis was performed using edgeR (Robinson et al., 2010; McCarthy et al., 2012). Data were first normalized using Counts Per Million (CPM) and examined by Multidimentional Scaling in the edgeR package (Robinson et al., 2010) to detect outliers. The data was then normalized using TMM (trimmed mean of M values) and comparisons were tested following recommendations in the User’s Guide. False discovery rates (FDR) were calculated using the Benjamini & Hochberg method (Benjamini & Hochberg, 1995).
Gene Ontology (GO) Term Enrichment Analysis
Gene symbols for differentially expressed genes were converted to entrez ids using biomaRt and analyzed with the clusterProfiler package in R (Durinck et al., 2009; Yu et al., 2012). A FDR cutoff of ≤0.05 was applied to focus the enrichment analysis on the significant genes. Gene ontology categories “Biological Processes”, “Cellular Components”, “Molecular Functions” and KEGG pathways were searched. Terms with FDR ≤0.05 were merged if more than 30% of genes overlapped.
Validation of RNAseq Data by qRT-PCR
Validation of RNA‐seq data for 3 different murine genes important for basal ganglia neurochemistry (Drd1, Penk, and Pdyn) was performed by qRT-PCR. The primers were TaqMan probes, where TaqMan assay primers and probes mix (assays-on-demand™ Gene expression Products) were obtained from (ABI, USA). The reaction mixture (20 µl) containing 1 µl of cDNA template, 1 µl each of primer and probe mix and (10 µl) TaqMan Fast Advanced PCR Master Mix (ABI, USA), brought to 20 uL with nuclease-free water. They were amplified with the following parameters, per manufacturer instructions: 50°C for 2 min, 95°C for 20 sec, and 40 cycles of 95°C for 1 sec and 60°C for 20 sec.
Cytokine Multiplex Assay
Frontal lobe tissue was prepared as specified by the Milliplex Multiplex Luminex assay manufacturer instructions. Specifically, 100 µg of protein was assessed for changes in pro-inflammatory cytokines using the Mouse High Sensitivity T Cell Milliplex Multiplex Luminexassay (Millipore Sigma, Merck KGaA, Darmstadt, Germany), following manufacturer instructions. Samples were analyzed by and data were collected from a Bio-Plex 200 system (Bio-Rad) and analyzed with Bio-Plex Manager software (Bio-Rad). Standard, sample, and control wells with bead counts <37 were excluded. The analysis software employed a five parameter logistic regression standard curve to determine sample cytokine levels.
Statistical Analysis
Sample processing was performed blind and the code denoting treatment groups was only provided for data analysis. The sample size of 7 was previously determined by the National Toxicology Program and from prior reports (King-Herbert & Thayer, 2006; Nayak et al., 2018). The ROUT method in GraphPad Prism was used to identify outliers, Q = 1. The Levine test was used to assess the homogeneity of variance, where p > 0.05 indicated homogeneity. Data were analyzed by one-way analysis of variance (ANOVA) using GraphPad Prism (GraphPad Prism, San Diego, CA, USA) when the variance was homogenous. When the assumption of homogeneity of variance was not met, the Kruskal-Wallis test was used, followed by post tests. Mean differences were analyzed by Bonferroni’s post-hoc analysis. Data are expressed as the mean ± SEM. A p-value <0.05 was considered statistically significant. When appropriate, representative images are shown.
Results
Short Term Aspergillus versicolor Exposure Is Sufficient to Trigger Neuroinflammation Gene Expression
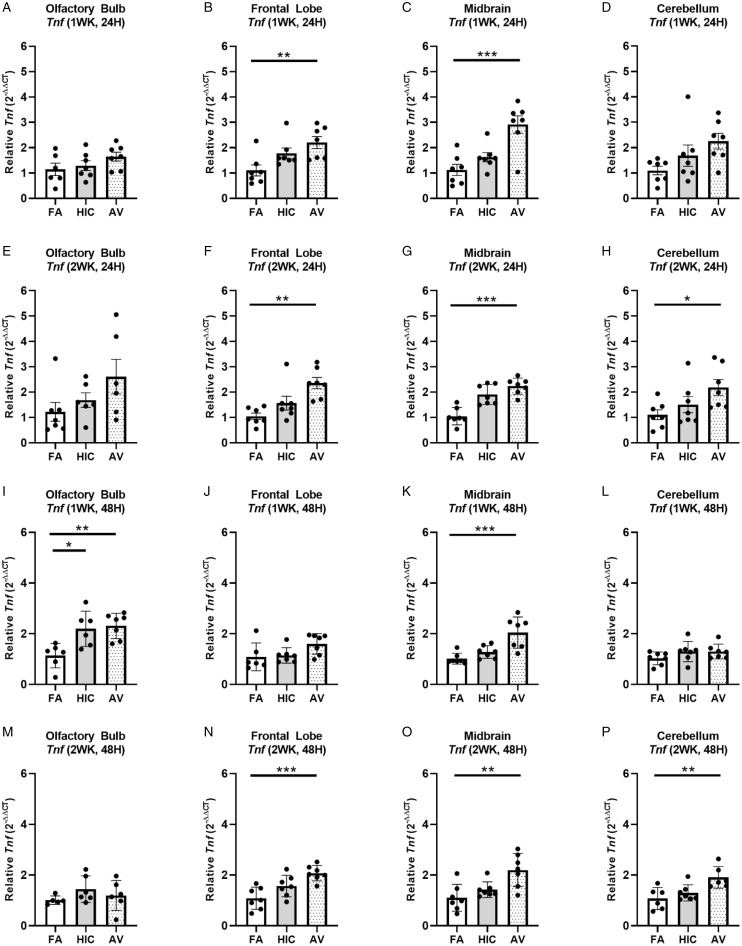
To begin to assess whether short term inhalation exposure to A. versicolor (twice a week, for either 1 or 2 weeks) was sufficient to trigger a neuroimmune response in the brain (olfactory bulb, frontal lobe, midbrain, and cerebellum), we first assessed whether the gene expression of the prototypical pro-inflammatory cytokine in the brain, Tnf, was modified when compared to mice exposed to filtered air. At 24H after the last exposure (Figure 2A to H), TnfmRNA was elevated in response to only viable A. versicolor in the frontal lobe (F(2,18)=6.294, p = 0.0074) (Figure 2B) and the midbrain (F(2,18) = 12.82, p = 0.0003) (Figure 2C), when the exposure duration was 1 week. The 2-week exposure resulted in an elevated Tnf mRNA response to only viable A. versicolor in the frontal lobe (F(2,18) = 9.524, p = 0.0012) (Figure 2F), the midbrain (F(2,18) = 21.19, p = 0.0001) (Figure 2G), and an additional region, the cerebellum (F(2,18) = 3.737, p = 0.0437) (Figure 2H). Importantly, the neuroimmune response generally persisted at 48H after the last A. versicolor exposure (Figure 2I to P), where the 48H post inhalation response for the 2-week exposure showed that only the viable A. versicolor triggered neuroinflammation gene expression and demonstrated the same pattern of elevated Tnf mRNA expression in the frontal lobe (F(2,18) = 11.02, p = 0.0005) (Figure 2N), the midbrain (F(2,18) = 8.265, p = 0.0028) (Figure 2O), and the cerebellum (F(2,16) = 7.267, p = 0.0063) (Figure 2P). For the 1-week exposure, the midbrain showed viable A. versicolor-induced elevation of Tnf gene expression after 48H (F(2,18) = 12.44, p = 0.0004) (Figure 2K). Interestingly, for the 1 week exposure at 48H after the last exposure, the olfactory bulb showed an elevated Tnf response to both the HIC and viable A. versicolor exposure, suggesting that this early short term response in olfactory bulb does not require viable filamentous fungi spores (F(2,16) = 8.129, p = 0.0055) (Figure 2I). Together, these data support that short term A. versicolor exposure at 1 and 2 weeks is sufficient to trigger a low grade neuroinflammation gene expression response that generally persists until at least 48H after the last exposure, where the midbrain and the frontal lobe show the most consistent elevated responses across all time points.
Figure 2.
Short term Aspergillus versicolor exposure elevates brain Tnf mRNA expression. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 1 (A–D) or 2 (E–H) weeks and Tnf mRNA expression was assessed 24H after the last exposure in several brain regions. At 48 H after the 1 (I–L) or 2 (M–P) week exposure, Tnf mRNA expression was assessed in several brain regions. Relative Tnf mRNA levels in the olfactory bulb (A, E, I, M), frontal lobe (B, F, J, N), midbrain (C, G, K, O), and cerebellum (G, H, L, P) were determined by qRT-PCR. Individual data points for an experimental animal are represented as black dots. Values were normalized to Gapdh using the 2-ΔΔCT method and are the mean ± SEM. *p < .05; **p < .01; ***p < .001 vs. filtered air control; n = 6–7.
4-Week Aspergillus versicolor Inhalation Elevates Neuroinflammation Gene Expression Without Loss of Hippocampal Neurogenesis
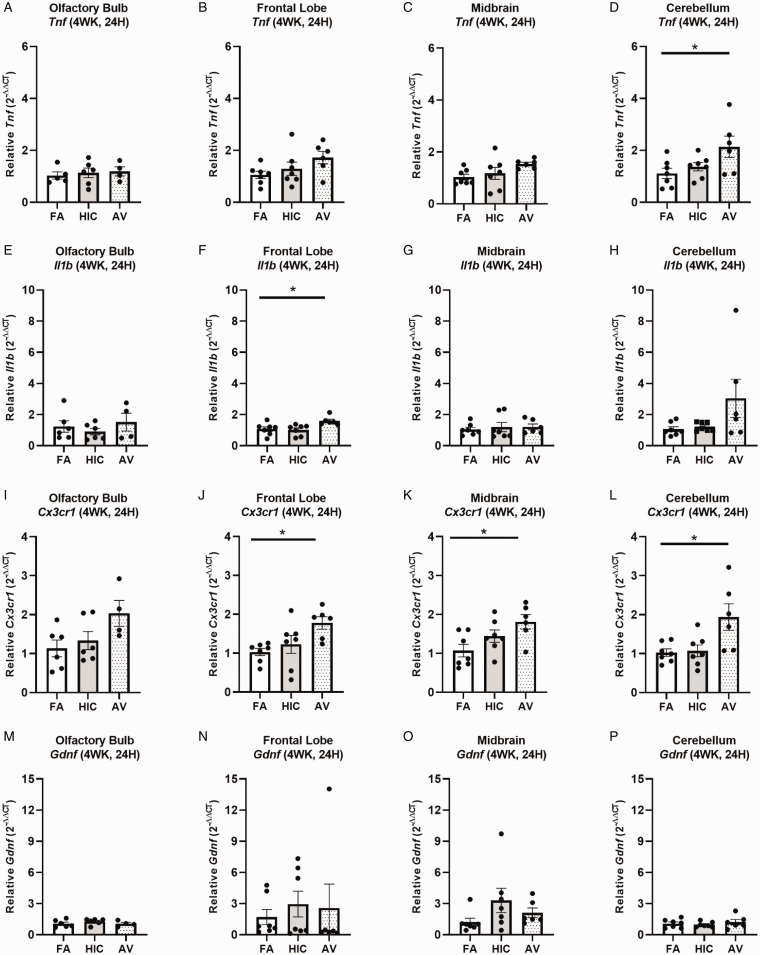
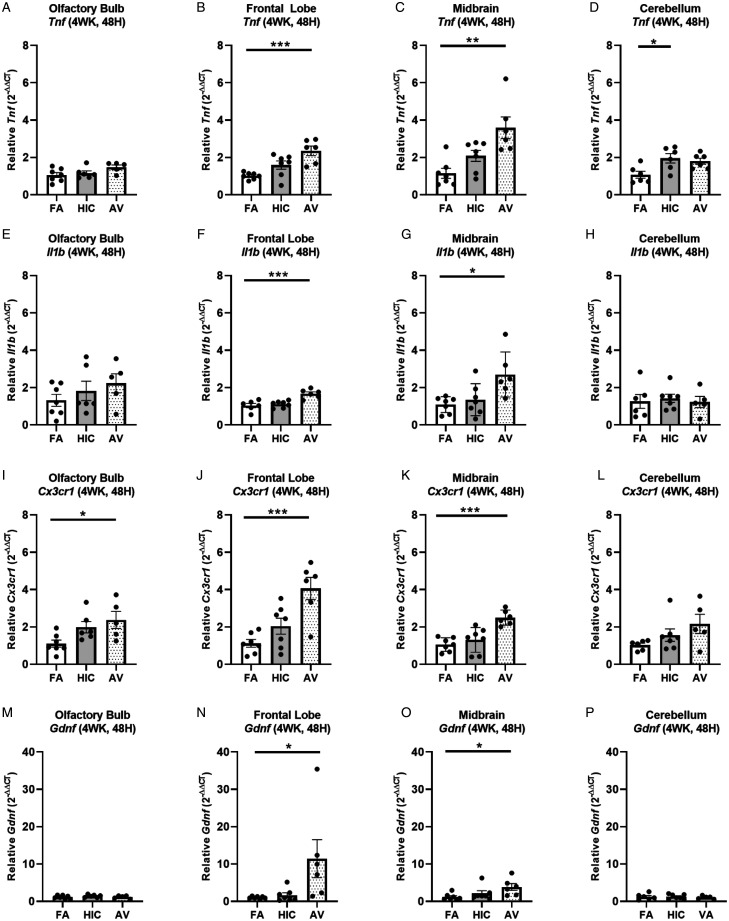
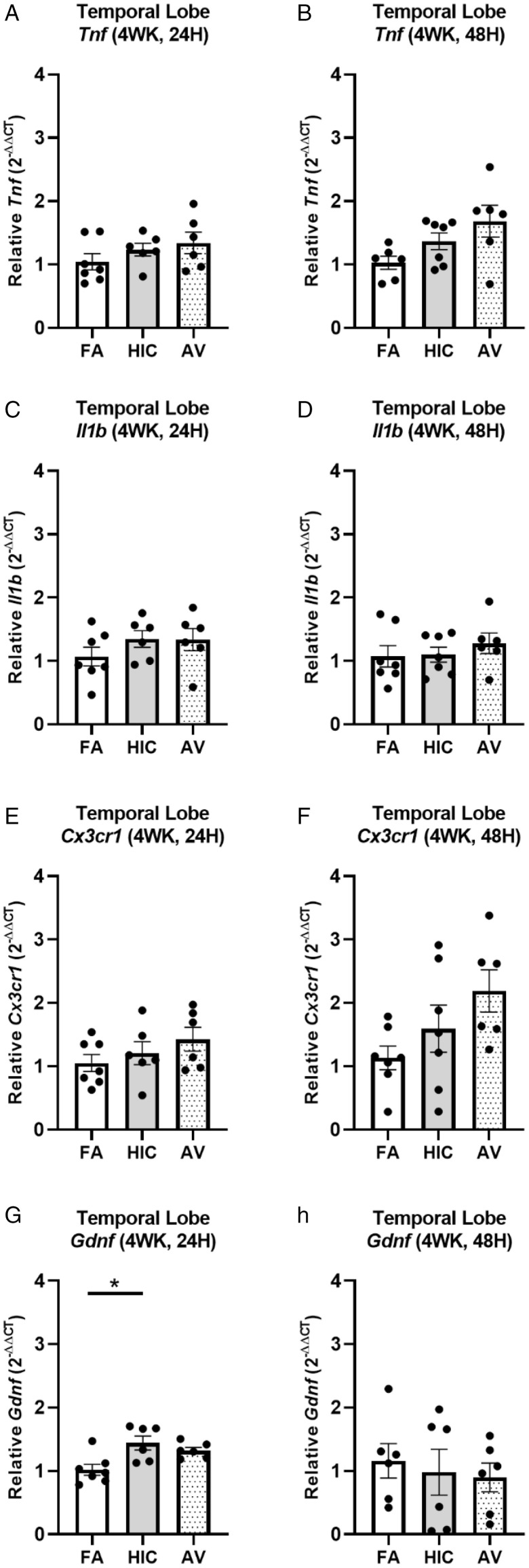
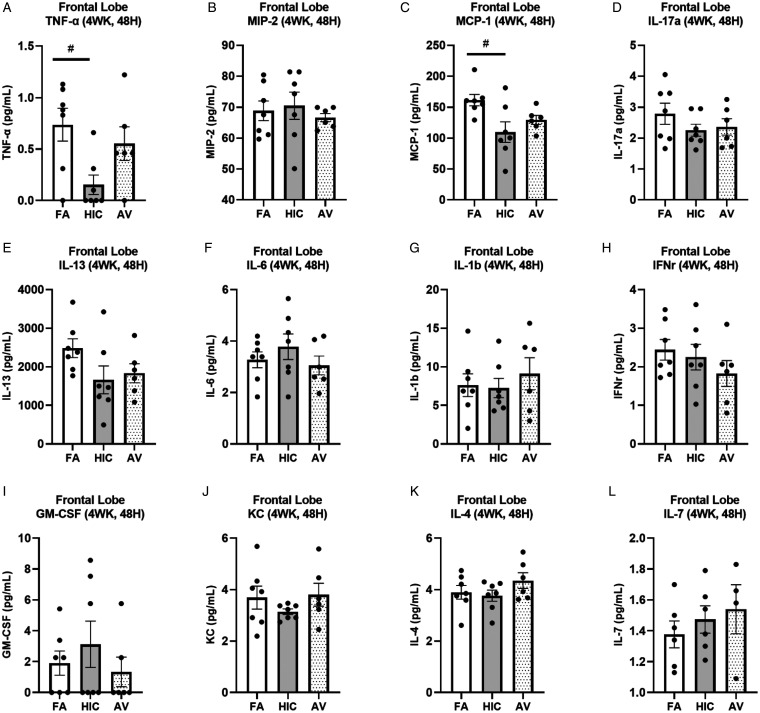
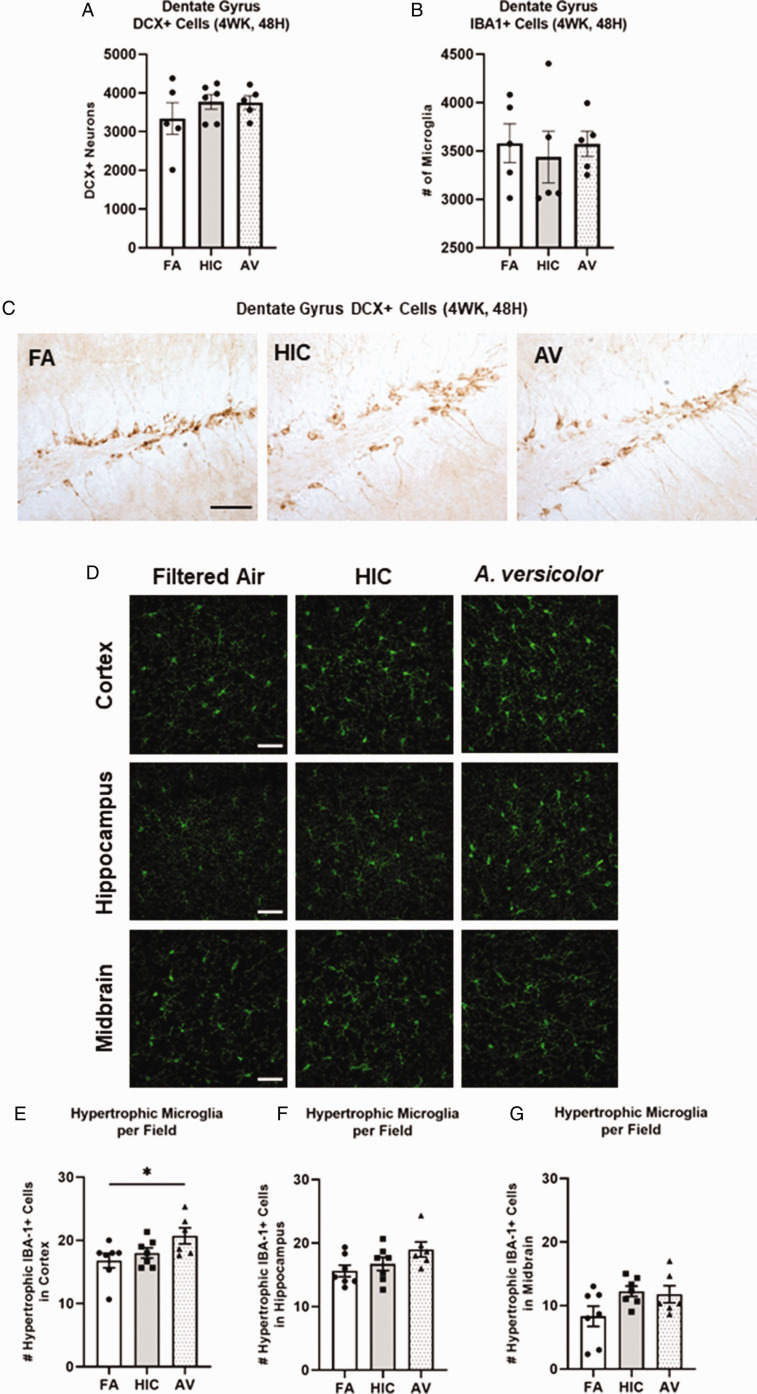
We next sought to assess how a longer A. versicolor exposure (twice a week for 4 weeks) would affect the neuroinflammation response and the potential impact on vulnerable neuronal populations in the hippocampus. At 24H after the final exposure (Figure 3A to P), significant increases in Cx3cr1 expression (frontal lobe (F(2,17) = 4.883, p = 0.0220) (Figure 3J), midbrain (F(2,17) = 4.698, p = 0.0212) (Figure 3K), and cerebellum (H(2,17) = 6.545, p = 0.0379) (Figure 3L) and Il1b expression in the frontal lobe (F(2,17) = 5.343, p = 0.0449) (Figure 3F) indicated viable A. versicolor triggered neuroinflammation. Tnf gene expression was only significantly upregulated in the cerebellum 24 H after the exposure F(2,17) = 3.892, p = 0.0450 (Figure 3D). On the tissue collected 48H after the last 4-week A. versicolor exposure (Figure 4A to P),the midbrain exhibited a consistent neuroinflammation gene expression response from only the viable A. versicolor exposure as evidenced by an increase in the expression of several genes: Tnf (F(2,17) = 9.739, p = 0.0012) (Figure 4C), Il1b (F(2,17) = 6.286, p = 0.0116) (Figure 4G), Cx3cr1 (F(2,17) = 15.05, p = 0.0002) (Figure 4K), and Gdnf (F(2,17) = 3.796, p = 0.0426) (Figure 4O). The frontal lobe at 48H post exposure also demonstrated a significant upregulation in response to only viable A. versicolor exposure in several biomarkers of neuroinflammation: Tnf (H(2,17) = 10.070, p = 0.0065) (Figure 4B), Cx3cr1 (F(2,17) = 12.25, p = 0.0004) (Figure 4J), (Il1b F(2,16) = 14.01, p = 0.0006) (Figure 4F), and Gdnf (F(2,17) = 4.552, p = 0.0437) (Figure 4N). At 48H after the last 4-week viable A. versicolor exposure, the olfactory bulb exhibited an upregulation of only Cx3cr1expression (F(2,15) = 4.606, p = 0.0342) (Figure 4I). Consistent with prior patterns seen in response to models of urban air pollution (Levesque et al., 2011a, 2011b), the cerebellum showed no significant pro-inflammatory response to viable A. versicolor at 48H after exposure, but a small Tnf response was significant in the HIC A. versicolor (F(2,15) = 5.445, p = 0.0227) (Figure 4D). Notably, no pro-inflammatory nor glial markers were upregulated in the temporal lobe at any time point after exposure (Figure 5A to H), p > 0.05). Analysis of protein collected from the frontal lobe failed to show any significant elevation of pro-inflammatory protein present (Figure 6A to L, p > 0.05). In addition, cell counts indicated that the number of microglial cells did not change in the hippocampus, further supporting that this was a mild neuroinflammation response (Figure 7B, p > 0.05). Cell counts of doublecortin positive (DCX+) cells in the hippocampus failed to show any significant differences in any exposure or time point, suggesting that this 4-week A. versicolor exposure has no impact on neurogenesis (Figure 7A and C, p > 0.05). However, changes in microglia morphology determined by increases in cell volume were significantly increased in the cortex, but the not the midbrain nor the hippocampus (Figure 7D–G). Taken together, these findings support that 4 weeks of exposure to viable A. versicolor triggers mild changes in pro-inflammatory gene transcription that is: the most consistent at 48H after the exposure stops, not translated to detectable changes in pro-inflammatory cytokine protein, and fails to cause toxicity in developing neurons in the hippocampus.
Figure 3.
Four-week Aspergillus versicolor exposure elevates neuroinflammation 24 hours after the final exposure. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolorconidia(HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks and markers of neuroinflammation were assessed in the olfactory bulb, frontal lobe, midbrain, and cerebellum 24H after the last exposure. Relative Tnf (A–D), IL1b (E–H), Cx3cr1 (I–L), and Gdnf (M–P) mRNA levels in the olfactory bulb, frontal lobe, midbrain, and cerebellum were determined using qRT-PCR. Individual data points for an experimental animal are represented as black dots. Values were normalized to Gapdh using the 2-ΔΔCT method and are the mean ± SEM. *p<.05 vs. filtered air control; n = 6–7.
Figure 4.
Four-week Aspergillus versicolor exposure elevates neuroinflammation gene expression 48 hours after the final exposure. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks and markers of neuroinflammation were assessed in the olfactory bulb, frontal lobe, midbrain, and cerebellum 48H after the last exposure. Relative Tnf (A–D), IL1b (E–H), Cx3cr1 (I–L), and Gdnf (M–P) mRNA levels in the olfactory bulb, frontal lobe, midbrain, and cerebellum were determined using qRT-PCR. Individual data points for an experimental animal are represented as black dots. Values were normalized to Gapdh using the 2-ΔΔCT method and are the mean ± SEM. *p < .05; **p < .01; ***p < .001 vs. filtered air control; n = 6–7.
Figure 5.
Four-week Aspergillus versicolor exposure fails to elevate neuroinflammation gene expression in the temporal lobe. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks and markers of neuroinflammation were assessed in the temporal lobe at 24H or at 48H after the last exposure. Relative Tnf (A and B), IL1b (C and D), Cx3cr1 (E and F), and Gdnf (G and H) mRNA levels in the temporal lobe were determined using qRT-PCR. Individual data points for an experimental animal are represented as black dots. Values were normalized to Gapdh using the 2-ΔΔCT method and are the mean ± SEM. *p < .05 vs. filtered air control; n = 6–7.
Figure 6.
Four-week Aspergillus versicolor exposure fails to elevate pro-inflammatory proteins in the frontal lobe 48 hours after the final exposure. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks and pro-inflammatory cytokines in the frontal lobe were assessed with a cytokine multiplex assay for changes in 48H after the last exposure. Relative pro-inflammatory cytokines levels (A–L) in the frontal lobe were determined using multiplex assay. Individual data points for an experimental animal are represented as black dots. The mean ± SEM. *p < .05 vs. filtered air control; n = 6–7.
Figure 7.
Four-week exposure to Aspergillus versicolor changes microglia morphology in the cortex, fails to affect morphology in other regions, and does not impact DCX+ neuron number. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (Air), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia(HIC), or 3 × 105 spores of viable Aspergillus versicolor twice weekly for 4 weeks. The number of DCX+ neurons (A) and IBA1+ microglia (B) in the dentate gyrus were counted with unbiased stereology. Representative images at 40× are shown of DCX+ neurons in the dentate gyrus of the hippocampus (C). Representative images of 40× images of IBA1+ (green) changes in microglia morphology in the CA1 region of the hippocampus, substantial nigra of the midbrain, and cortex are depicted (D). The number of hypertrophic IBA1+ microglia cells were counted in the cortex (E), and hippocampus (F), and the midbrain (G). Hypertrophic cell = volume >500 µm3. The scale bar indicates 50 microns. Individual data points for an experimental animal are represented as black dots. *p < .05 vs. filtered air control; † p < .05 vs. heat-inactivated control. n = 6–7.
4-Week Aspergillus versicolor Inhalation Results in a Unique Midbrain Transcriptomic Signature
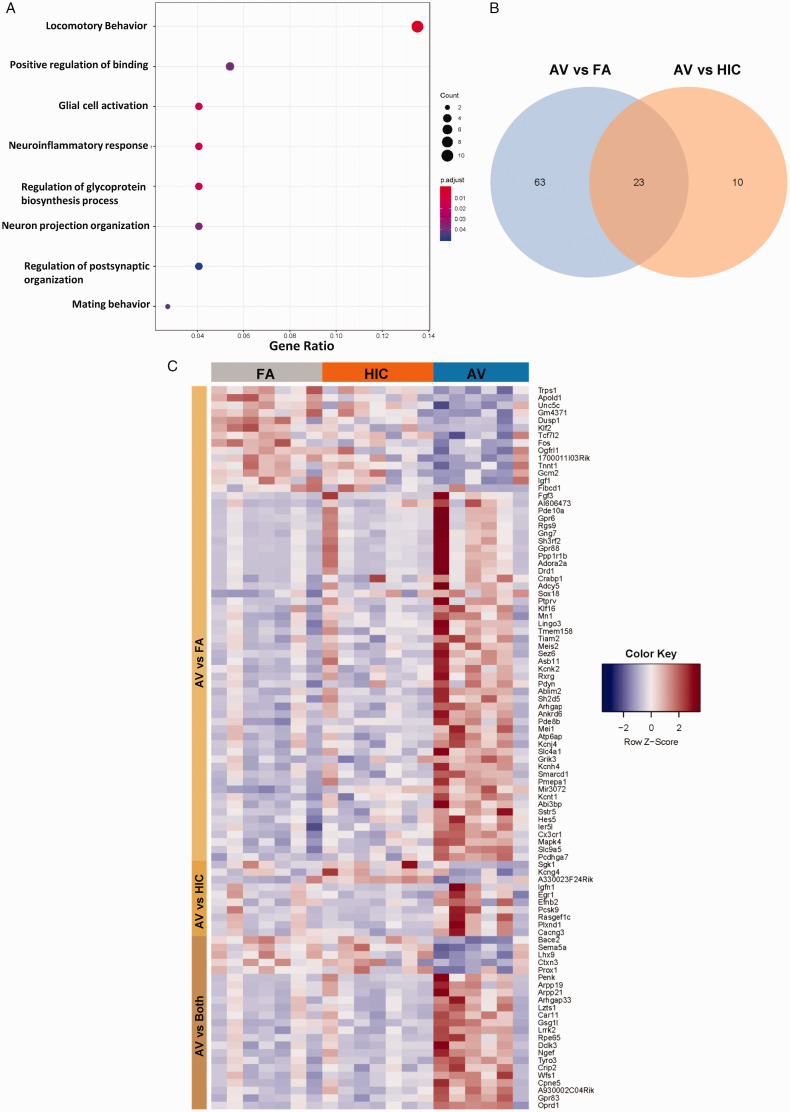
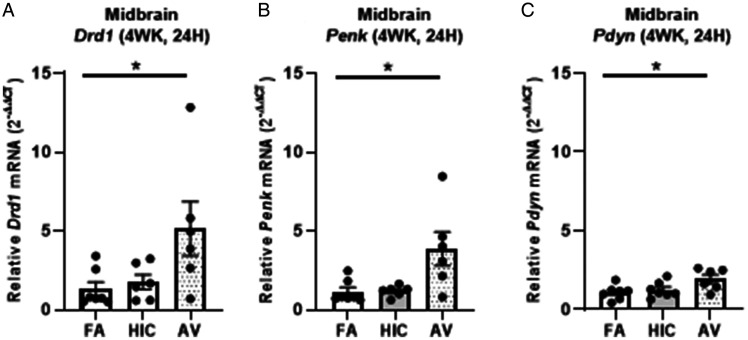
Additional inquiry sought to explore the transcriptional changes elicited by A. versicolor in the region exhibiting the most robust response, the midbrain 48H after the last 4-weekA. versicolor exposure. Bulk RNAseq analysis of the 48H midbrain tissue revealed a total of 86 genes significantly differentially expressed following A. versicolor exposure compared to the filtered air control group (Figure 8; Table 3, p < 0.05), where 23 of these genes were also expressed following HIC A. versicolor exposure(Figure 8B, p < 0.05), demonstrating some response following the inhalation of inactive spores. GO analysis revealed that A. versicolor significantly enriched gene expression in key biological pathways of interest, including neuroinflammatory response and glial cell activation (Figure 8A, p < 0.05). Notably, Cxc3r1, which was previously determined by qRT-PCR to be upregulated in several tissues and time points in response to viable A. versicolor was confirmed to be significantly upregulated 1.9 - fold by RNA-seq analysis (Table 3). GO analysis highlighted gene expression in the locomotor behavior as the most abundant group changed, consistent with the fact the midbrain contains the substantia nigra, a key region of the basal ganglia that is critical for regulating motor behavior. Hierarchical clustering of the differentially expressed genes revealed clear changes in the overall gene expression pattern following viable A. versicolor exposure compared to control (Figure 8C, p < 0.05). The genes significantly modified following viable A. versicolor exposure after correction are listed in supplemental Table 3. A closer look at genes with the highest fold change in the midbrain (Table 3, p < 0.05) in response to viable A. versicolor exposure revealed a list of receptors and signaling pathways associated with neurotransmitter signaling the basal ganglia. qRT-PCR analysis confirmed elevation of midbrain Drd1(Dopamine Receptor D1), Penk (Proenkephalin-A), and Pdyn (Prodynorphin) gene expression (F(2,16) = 4.265, p = 0.0455) (Figure 9A), (H(2, 16) = 7.781, p = 0.0104) (Figure 9B), and (F(2, 17) = 4.194, p = 0.0400) (Figure 9C), supporting that that A. versicolor exposure may dysregulate basal ganglia neurotransmitter receptors and neuropeptides.
Figure 8.
Four-week Aspergillus versicolor exposure results in a unique transcriptional signature in midbrain tissue. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks and bulk RNA-seq analysis was performed on midbrain tissue 48H after the last exposure. Significantly enriched biological pathways were identified by GO analysis. Gene Ratio refers to the number of significant genes matched to the term/total number of significant genes (A), a Venn diagram illustrates the overlap of significantly modified genes by exposure (B), and hierarchical clustering of the differentially expressed genes, using the RNA‐seq data derived from the three exposures was assessed.
Table 3.
Genes Significantly Modified by 4 Week A. versicolor Exposure Compared to Filtered Air.
| Gene | Full name | Fold change | FDR |
|---|---|---|---|
| Adora2a | adenosine A2a receptor | 10.9472837 | 0.000415 |
| Ptprv | protein tyrosine phosphatase, receptor type, V | 9.013871799 | 0.003516 |
| Sh3rf2 | SH3 domain containing ring finger 2 | 7.875241279 | 0.030416 |
| AI606473 | expressed sequence AI606473 | 6.498195103 | 0.019815 |
| Rpe65 | retinal pigment epithelium 65 | 4.559200812 | 0.022028 |
| Drd1 | dopamine receptor D1 | 4.083336682 | 0.002994 |
| Ankrd63 | ankyrin repeat domain 63 | 4.080625751 | 0.006816 |
| Mei1 | meiotic double-stranded break formation protein 1 | 4.014669637 | 0.030416 |
| Gpr6 | G protein-coupled receptor 6 | 3.910378715 | 0.005819 |
| Gpr88 | G-protein coupled receptor 88 | 3.729838181 | 0.004266 |
| Asb11 | ankyrin repeat and SOCS box-containing 11 | 3.63581009 | 0.004324 |
| Penk | preproenkephalin | 3.587034698 | 0.000101 |
| A930002C04Rik | RIKEN cDNA A930002C04 gene | 3.584852141 | 0.00498 |
| Ppp1r1b | protein phosphatase 1, regulatory inhibitor subunit 1B | 3.559500978 | 0.008428 |
| Fgf3 | fibroblast growth factor 3 | 3.34480696 | 0.036596 |
| Kcnh4 | potassium voltage-gated channel, subfamily H (eag-related), member 4 | 3.112689051 | 0.003664 |
| Abi3bp | ABI gene family, member 3 (NESH) binding protein | 2.948697477 | 0.004324 |
| Atp6ap1l | ATPase, H+ transporting, lysosomal accessory protein 1-like | 2.554556989 | 0.008428 |
| Rgs9 | regulator of G-protein signaling 9 | 2.488406437 | 0.004324 |
| Meis2 | Meis homeobox 2 | 2.482523431 | 0.00498 |
| Kcnj4 | potassium inwardly-rectifying channel, subfamily J, member 4 | 2.456610173 | 0.012027 |
| Crabp1 | cellular retinoic acid binding protein I | 2.454263194 | 0.022716 |
| Cpne5 | copine V | 2.378022974 | 0.000101 |
| Slc4a11 | solute carrier family 4, sodium bicarbonate transporter-like, member 11 | 2.211832227 | 0.009963 |
| Ngef | neuronal guanine nucleotide exchange factor | 2.076791901 | 1.93E-05 |
| Sstr5 | somatostatin receptor 5 | 2.060885658 | 0.032776 |
| Rxrg | retinoid X receptor gamma | 1.996022823 | 0.008452 |
| Arpp21 | cyclic AMP-regulated phosphoprotein, 21 | 1.986804883 | 0.014142 |
| Arhgap10 | Rho GTPase activating protein 10 | 1.966255124 | 0.023956 |
| Gng7 | guanine nucleotide binding protein (G protein), gamma 7 | 1.92922481 | 0.02271 |
| Wfs1 | wolframin ER transmembrane glycoprotein | 1.921515273 | 0.000357 |
| Pdyn | prodynorphin | 1.913202775 | 0.01699 |
| Pde10a | phosphodiesterase 10A | 1.906578023 | 0.030416 |
| Lzts1 | leucine zipper, putative tumor suppressor 1 | 1.840120478 | 0.018402 |
| Gpr83 | G protein-coupled receptor 83 | 1.799091412 | 0.000195 |
| Hes5 | hes family bHLH transcription factor 5 | 1.786780965 | 0.013532 |
| Tmem158 | transmembrane protein 158 | 1.743780225 | 0.035381 |
| Oprd1 | opioid receptor, delta 1 | 1.687927309 | 0.000101 |
| Tiam2 | T cell lymphoma invasion and metastasis 2 | 1.678126517 | 0.03842 |
| Lingo3 | leucine rich repeat and Ig domain containing 3 | 1.661601028 | 0.01095 |
| Arhgap33 | Rho GTPase activating protein 33 | 1.6561737 | 0.013532 |
| Mir3072 | microRNA 3072 | 1.64422367 | 0.020116 |
| Arpp19 | cAMP-regulated phosphoprotein 19 | 1.640835825 | 0.012022 |
| Mn1 | meningioma 1 | 1.636881969 | 0.004324 |
| Tyro3 | TYRO3 protein tyrosine kinase 3 | 1.594697407 | 0.002743 |
| Gsg1l | GSG1-like | 1.573627024 | 0.006816 |
| Adcy5 | adenylate cyclase 5 | 1.528203477 | 0.047749 |
| Klf16 | Kruppel-like factor 16 | 1.516215729 | 0.043358 |
| Dclk3 | doublecortin-like kinase 3 | 1.507508487 | 0.033981 |
| Sox18 | SRY (sex determining region Y)-box 18 | 1.42519843 | 0.036965 |
| Kcnk2 | potassium channel, subfamily K, member 2 | 1.41812487 | 0.025752 |
| Ier5l | immediate early response 5-like | 1.415408171 | 0.037193 |
| Lrrk2 | leucine-rich repeat kinase 2 | 1.37322213 | 0.014676 |
| Pmepa1 | prostate transmembrane protein, androgen induced 1 | 1.364965461 | 0.000415 |
| Sh2d5 | SH2 domain containing 5 | 1.358951454 | 0.014676 |
| Crip2 | cysteine rich protein 2 | 1.348787861 | 0.023956 |
| Ablim2 | actin-binding LIM protein 2 | 1.344357391 | 0.014142 |
| Car11 | carbonic anhydrase 11 | 1.339516476 | 0.012996 |
| Sez6 | seizure related gene 6 | 1.328336163 | 0.026462 |
| Pcdhga7 | protocadherin gamma subfamily A, 7 | 1.312608893 | 0.009963 |
| Slc9a5 | solute carrier family 9 (sodium/hydrogen exchanger), member 5 | 1.311851384 | 0.004021 |
| Grik3 | glutamate receptor, ionotropic, kainate 3 | 1.302810927 | 0.019477 |
| Kcnt1 | potassium channel, subfamily T, member 1 | 1.302045508 | 0.003154 |
| Mapk4 | mitogen-activated protein kinase 4 | 1.295745705 | 0.005572 |
| Cx3cr1 | chemokine (C-X3-C motif) receptor 1 | 1.295376554 | 0.002993 |
| Smarcd1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | 1.278578756 | 0.016504 |
| Pde8b | phosphodiesterase 8B | 1.26565131 | 0.025752 |
| Trps1 | transcriptional repressor GATA binding 1 | –1.26429562 | 0.033981 |
| Ogfrl1 | opioid growth factor receptor-like 1 | –1.30158073 | 0.035836 |
| Unc5c | unc-5 netrin receptor C | –1.35859396 | 0.005941 |
| Sema5a | semaphorin5A | –1.39986876 | 0.013532 |
| Fos | FBJ osteosarcoma oncogene | –1.5146754 | 0.002993 |
| Igf1 | insulin-like growth factor 1 | –1.51550774 | 0.023956 |
| Klf2 | Kruppel-like factor 2 (lung) | –1.54770363 | 0.000972 |
| Prox1 | prospero homeobox 1 | –1.55096535 | 0.019615 |
| Dusp1 | dual specificity phosphatase 1 | –1.56284447 | 0.000122 |
| Bace2 | beta-site APP-cleaving enzyme 2 | –1.62516347 | 0.001442 |
| Tnnt1 | troponin T1, skeletal, slow | –1.66510921 | 0.015881 |
| Tcf7l2 | transcription factor 7 like 2, T cell specific, HMG box | –1.76434402 | 0.019615 |
| Apold1 | apolipoprotein L domain containing 1 | –1.85430711 | 0.000101 |
| Lhx9 | LIM homeobox protein 9 | –1.95702439 | 0.000972 |
| Fibcd1 | fibrinogen C domain containing 1 | –2.33005196 | 0.00701 |
| Ctxn3 | cortexin 3 | –2.65994007 | 0.008139 |
| Gm4371 | eukaryotic translation initiation factor 3, subunit I pseudogene | –2.85369148 | 0.005819 |
| 1700011I03Rik | NA | –2.91923417 | 0.019477 |
| Gcm2 | glial cells missing homolog 2 | –10.0585016 | 0.010923 |
Figure 9.
Four-week Aspergillus versicolor exposure elevates basal ganglia neurotransmission genes in the midbrain. Eight week-old female B6C3F1/N mice were exposed in a nose only chamber to filtered air (FA), 3 × 105 spores of heat-inactivated Aspergillus versicolor conidia (HIC), or 3 × 105 spores of viable Aspergillus versicolor (AV) twice weekly, for 4 weeks and markers of neuroinflammation were assessed in the midbrain 48H after the last exposure. Relative Drd1 (A), Penk (B), and PDyn (C) mRNA levels in the midbrain were determined using qRT-PCR. Individual data points for an experimental animal are represented as black dots. Values were normalized to Gapdh using the 2-ΔΔCT method and are the mean ± SEM. *p<.05 vs. filtered air control; n = 6–7.
Discussion
Increasing evidence points to a role for inhaled indoor pollutants and fungal bioaerosols in CNS effects and behavior deficits, but the potential underlying mechanisms are poorly understood. Here, we demonstrate that a moderate A. versicolor exposure, the most common opportunistic filamentous fungal species in damp indoor environments, triggers neuroinflammation gene expression with short term exposures (1 and 2 weeks) in multiple brain regions. The frontal lobe and midbrain were more sensitive to the neuroimmune changes caused by a 4-week A. versicolor exposure, with the most robust and consistent elevation of pro-inflammatory and glial gene expression markers. Further transcriptomic inquiry into the midbrain response to 4 weeks of A. versicolor exposure revealed changes in signaling and neurotransmitter gene expression relevant to basal ganglia function. Together, these findings support that the brain detects and responds to A. versicolor inhalation with glial, pro-inflammatory, and neurochemistry gene expression changes, providing much needed insight into the CNS effects of filamentous fungi inhalation.
While neuroinflammation is implicated in the deleterious CNS effects of many CNS diseases and environmental toxicants, the presence of neuroinflammation itself may not necessarily be overtly neurotoxic (Jayaraj et al., 2017). This premise is supported in the current study, as pro-inflammatory and glial gene markers were elevated (Figures 2 to 4), without any evidence of cell loss in the hippocampus (Figure 7), two effects that occurred together with high Stachybotrys chartarum exposure in mice (Harding et al., 2020), indicating that the species and exposure level for fungi are important for CNS effects. Unexpectedly, Tnf was upregulated in the 2 (Figure 3) and 4 week (Figure 4) viable A. versicolor exposure in the cerebellum, a brain region with lower numbers of glial cells, a pattern different than seen with urban air pollution exposure, suggesting that A. versicolor may exhibit a more global CNS neuroimmune effect than some urban air pollution exposures. There were also no significant changes to microglia morphology or number in response to viable A. versicolor exposure in the hippocampus (Figure 7), but there was a significant morphology change in the cortex (Figure 7) further supporting that A. versicolor produces a low level neuroinflammation response. Often the HIC and AV were not significantly different, suggesting that perhaps there may be some subtle, but not significant effect of HIC.
Neuroinflammation triggered by an inhaled toxin or toxicant is a complex process with multiple pathways, including translocation of antigens or cytokines into the brain through the olfactory tubercles and in response to the systemic effects of pulmonary immune responses (Jayaraj et al., 2017). How A. versicolor inhalation elicits a neuroimmune response has yet to be tested, but information on the response time and the role of viability in the response may provide insight. The majority of the significant neuroimmune changes reported in this study required exposure to viable A. versicolor, suggesting that the initial presence of the antigen alone, at the levels present with this exposure, may be insufficient to trigger neuroimmune response in most brain regions and that a secondary response, perhaps to increasing secondary metabolites or the pulmonary immune response (Barnes et al., 2020), may be important. However, both the 1-week viable and HIC A. versicolor exposure triggered Tnf mRNA expression in the olfactory bulb 48H after exposure (Figure 2), indicating that the initial response to antigens present on both viable and HIC A. versicolor may be sufficient for the olfactory bulb effect. The only other neuroimmune effect of HIC A. versicolor exposure occurred with the 4-week exposure in the temporal lobe 24H after exposure (Figure 4), when compared to the other brain regions, emphasizing that a response to HIC A. versicolor was rare. In addition, the largest and most consistent Tnf response and neuroimmune changes occurred in the midbrain 48H after the last 4-week viable A. versicolor exposure (Figure 4), supporting an accumulation of the A. versicolor neuroinflammation gene expression response over repeated exposures and time post exposure. As such, these data suggest that inhalation of viable A. versicolor triggers a modest, but significant, sterile neuroimmune transcriptional response that may be predominantly associated with secondary products generated. Future research will need to define the potential role of the immune mechanisms in the lung, infiltrating immune cells, and brain region specific pattern recognition receptors that might be driving these neuroimmune changes. It will also be critical to establish how long neuroimmune changes persist after exposure and the consequences for long term and lifetime exposures.
Despite early human studies (Baldo et al., 2002; Gordon et al., 2004; Lieberman et al., 2006; Ratnaseelan et al., 2018) and animal research studies (Harding et al., 2020), due to current lack of direct inquiry and mechanistic insight, debate remains on the CNS effects of filamentous fungi exposure. Given that patient (Baldo et al., 2002; Gordon et al., 2004; Lieberman et al., 2006; Ratnaseelan et al., 2018) and animal reports (Girman et al., 2002; Rea et al., 2003; Gordon et al., 2004; Harding et al., 2020) focused on cognitive effects, which implicates the frontal lobe and hippocampus, it was surprising that some of the most robust neuroimmune effects occurred in the midbrain, the region that houses the substantia nigra, which is important for motor function. However, this elevation of pro-inflammatory markers in the midbrain region is consistent with what has been seen with diesel exhaust (Levesque et al., 2011a, 2011b), an inhaled air pollutant. Further inquiry revealed that A. versicolor exposure dysregulated the expression of neurotransmission genes relevant to basal ganglia function (Figures 8 and 9; Table 3). More specifically, qRT-PCR confirmation of highly expressed genes identified by RNA-seq show that Drd1, the D1 dopamine subreceptor abundant in the basal ganglia, also increased expression in response to only viable A. versicolor exposure. Penk, a neuropeptide with instriatolateral-pallidal neurons, was upregulated in only viable A. versicolor exposure. In addition, Pdyn, another basal ganglia neuropeptide, was also discovered to be upregulated in response to only viable A. versicolor. These neuropeptides and the D1 receptor are important for motor behavior in the basal ganglia and are the first neurotransmission genes reported to show perturbed expression with inhaled fungal exposure. Interestingly, a previous report has implicated the common secondary fungal metabolite, semiochemical 1-octen-3-ol, in deficits in dopamine levels and dopamine neuron degeneration in Drosophila melanogaster, further supporting the potential for filamentous fungi to cause substantia nigra pathology (Inamdar et al., 2013). As such, these data support further inquiry with future studies into the impact on potential behavioral changes, the specific neurochemical changes caused by A. versicolor, and the surprising possible association with Parkinson’s disease, Huntington’s disease, and perhaps addiction phenotypes.
In summary, this work demonstrates that viable A. versicolor inhalation can cause neuroinflammation in multiple brain regions at both 24H and 48H after the final exposure, where the number of exposures over time will increase neuroinflammation gene expression in the midbrain and can persist for at least 48H after the last exposure. While there is an extremely low CNS transcriptional response to heat-inactivated A. versicolor that often fails to meet significance, we show that spore viability is important for most changes in neuroinflammation gene expression, potentially implicating the secondary products produced by viable filamentous fungi in the neuroimmune response. Notably, inhalation of viable A. versicolor caused a unique transcriptional CNS phenotype that includes upregulation of genes involved in the neuroimmune response, glial cell markers, and neurotransmission. Taken together, these data are the first to report that the brain detects and responds to moderate inhalation exposure of the common opportunistic filamentous fungi, A. versicolor, with parenchymal CNS changes. These findings provide much needed insight into how the allergic pulmonary response can affect the brain and highlight the future need to understand its potential impact on CNS function and disease.
Summary
In the current report, we demonstrate that a moderate exposure to live Aspergillus versicolor, a common indoor air pollutant in damp environments, causes a unique neuroimmune and neurotransmission transcriptional signature, highlighting a potential role for the allergic pulmonary immune response in neuroimmune regulation.
Abbreviations
Aspergillus (A.)
Central nervous system (CNS)
C-X3-C motif chemokine receptor 1 (Cxc3r1)
Glial cell line-derived neurotrophic factor (Gdnf)
Dopamine receptor D1 (Drd1)
Filtered Air (FA)
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
Heat inactivated conidia (HIC)
Interleukin 1 beta (IL-1β)
Ionized calcium-binding adapter molecule 1 (IBA-1)
Proenkephalin-A (Penk)
Prodynorphin (Pdyn)
Reactive oxygen species (ROS)
Tumor necrosis factor alpha (TNFα)
Supplemental Material
Supplemental material, sj-xlsx-1-asn-10.1177_17590914211019886 for Aspergillus versicolor Inhalation Triggers Neuroimmune, Glial, and Neuropeptide Transcriptional Changes by Thatcher B. Ladd, James A. Johnson Jr., Christen L. Mumaw, Hendrik J. Greve, Xiaoling Xuei, Ed Simpson, Mark A. Barnes, Brett J. Green, Tara L. Croston, Chandrama Ahmed, Angela Lemons, Donald H. Beezhold and Michelle L. Block in ASN Neuro
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the NIH R01 ES028104 (and the NIA supplement) and graciously supported by both the VA Merit Award I01 BX004161 and NIH RO1 ES029835GW awarded to Michelle L. Block. This study was also supported in part by NIOSH/CDC interagency agreement AES 12007001-1-0-6 and CDC intramural funds (927ZLCT). The findings and conclusions in this study are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention. The contents also do not represent the views of the U.S. Department of Veterans Affairs nor the United States Government.
ORCID iDs: Ed Simpson https://orcid.org/0000-0001-9015-9864
Angela Lemmons https://orcid.org/0000-0003-3057-9888
Michelle L. Block https://orcid.org/0000-0001-8303-7914
Supplemental Material: Supplemental material for this article is available online.
References
- Baldo J. V., Ahmad L., Ruff R. (2002). Neuropsychological performance of patients following mold exposure. Appl Neuropsychol, 9(4), 193–202. [DOI] [PubMed] [Google Scholar]
- Barnes M. A., Croston T. L., Lemons A. R., Rush R. E., Beezhold D. H., Green B. J. (2020). Subchronic inhalation of Aspergillus versicolor conidia leads to sustained innate immune responses and is associated with irreversible remodeling of pulmonary arterial tissue following recovery. J Immunol, 204. [Google Scholar]
- Beguin H., Nolard N. (1994). Mould biodiversity in homes I. Air and surface analysis of 130 dwellings. Aerobiologia, 10(2–3), 157–166. [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B, 57, 289–300. [Google Scholar]
- Breese M. R., Liu Y. (2013). NGSUtils: A software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics, 29(4), 494–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk A. D., Green B. J., Lemons A. R., Nayak A. P., Goldsmith W. T., Kashon M. L., Anderson S. E., Hettick J. M., Templeton S. P., Germolec D. R., Beezhold D. H. (2014). A murine inhalation model to characterize pulmonary exposure to dry Aspergillus fumigatus conidia. PLoS One, 9(10), e109855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Garcidueñas L., Kavanaugh M., Block M., D'Angiulli A., Delgado-Chávez R., Torres-Jardón R., González-Maciel A., Reynoso-Robles R., Osnaya N., Villarreal-Calderon R., Guo R., Hua Z., Zhu H., Perry G., Diaz P. (2012). Neuroinflammation, hyperphosphorylated tau, diffuse amyloid plaques, and down-regulation of the cellular prion protein in air pollution exposed children and young adults. J Alzheimer's Dis, 28(1), 93–107. [DOI] [PubMed] [Google Scholar]
- Calderón-Garcidueñas L., Reed W., Maronpot R. R., Henríquez-Roldán C., Delgado-Chavez R., Calderón-Garcidueñas A., Dragustinovis I., Franco-Lira M., Aragón-Flores M., Solt A. C., Altenburg M., Torres-Jardón R., Swenberg J. A. (2004). Brain inflammation and Alzheimer's-like pathology in individuals exposed to severe air pollution. Toxicol Pathol, 32(6), 650–658. [DOI] [PubMed] [Google Scholar]
- Calderón-Garcidueñas L., Solt A. C., Henríquez-Roldán C., Torres-Jardón R., Nuse B., Herritt L., Villarreal-Calderón R., Osnaya N., Stone I., García R., Brooks D. M., González-Maciel A., Reynoso-Robles R., Delgado-Chávez R., Reed W. (2008). Long-term air pollution exposure is associated with neuroinflammation, an altered innate immune response, disruption of the blood-brain barrier, ultrafine particulate deposition, and accumulation of amyloid beta-42 and alpha-synuclein in children and young adults. Toxicol Pathol, 36(2), 289–310. [DOI] [PubMed] [Google Scholar]
- Campbell A., Araujo J. A., Li H., Sioutas C., Kleinman M. (2009). Particulate matter induced enhancement of inflammatory markers in the brains of apolipoprotein E knockout mice. J Nanosci Nanotechnol, 9(8), 5099–5104. [DOI] [PubMed] [Google Scholar]
- Campbell A., Oldham M., Becaria A., Bondy S. C., Meacher D., Sioutas C., Misra C., Mendez L. B., Kleinman M. (2005). Particulate matter in polluted air may increase biomarkers of inflammation in mouse brain. Neurotoxicology, 26(1), 133–140. [DOI] [PubMed] [Google Scholar]
- Costa L. G., Cole T. B., Coburn J., Chang Y. C., Dao K., Roque P. J. (2017). Neurotoxicity of traffic-related air pollution. Neurotoxicology, 59, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston T. L., Lemons A. R., Barnes M. A., Goldsmith W. T., Orandle M. S., Nayak A. P., Germolec D. R., Green B. J., Beezhold D. H. (2020). Inhalation of Stachybotrys chartarum fragments induces pulmonary arterial remodeling. Am J Respir Cell Mol Biol, 62(5), 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., Huber W. (2009). Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat Protoc, 4(8), 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzel R., Rylander R. (1999). Indoor mold and children's health. Environ Health Perspect, 107(Suppl 3), 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girman , J. R., Baker B. J., Burton L. E. (2002). Prevalence of potential sources of indoor air pollution in U.S. Office Buildings. In Proceedings of Indoor Air 2002, Vol. IV, pp. 438–443.
- Gordon W. A., Cantor J. B., Johanning E., Charatz H. J., Ashman T. A., Breeze J. L., Haddad L., Abramowitz S. (2004). Cognitive impairment associated with toxigenic fungal exposure: A replication and extension of previous findings. Appl Neuropsychol, 11(2), 65–74. [DOI] [PubMed] [Google Scholar]
- Harding C. F., Pytte C. L., Page K. G., Ryberg K. J., Normand E., Remigio G. J., DeStefano R. A., Morris D. B., Voronina J., Lopez A., Stalbow L. A., Williams E. P., Abreu N. (2020). Mold inhalation causes innate immune activation, neural, cognitive and emotional dysfunction. Brain Behav Immun, 87, 218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heseltine E., Rosen J., & World Health Organization. (2009). WHO guidelines for indoor air quality: Dampness and mould. WHO: Copenhagen. [PubMed] [Google Scholar]
- Hutson C. B., Lazo C. R., Mortazavi F., Giza C. C., Hovda D., Chesselet M. F. (2011). Traumatic brain injury in adult rats causes progressive nigrostriatal dopaminergic cell loss and enhanced vulnerability to the pesticide paraquat. J Neurotrauma, 28(9), 1783–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvonen S., Lohi J., Tuuminen T. (2020). Moist and mold exposure is associated with high prevalence of neurological symptoms and MCS in a Finnish hospital workers cohort. Saf Health Work, 11(2), 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamdar A. A., Hossain M. M., Bernstein A. I., Miller G. W., Richardson J. R., Bennett J. W. (2013). Fungal-derived semiochemical 1-octen-3-ol disrupts dopamine packaging and causes neurodegeneration. Proc Natl Acad Sci U S A, 110(48), 19561–19566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraj R. L., Rodriguez E. A., Wang Y., Block M. L. (2017). Outdoor ambient air pollution and neurodegenerative diseases: The neuroinflammation hypothesis. Curr Environ Health Rep, 4(2), 166–179. [DOI] [PubMed] [Google Scholar]
- King-Herbert A., Thayer K. (2006). NTP workshop: Animal models for the NTP rodent cancer bioassay: Stocks and strains – Should we switch? Toxicol Pathol, 34(6), 802–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepeis N. E., Nelson W. C., Ott W. R., Robinson J. P., Tsang A. M., Switzer P., Behar J. V., Hern S. C., Engelmann W. H. (2001). The national human activity pattern survey (NHAPS): A resource for assessing exposure to environmental pollutants. J Expo Anal Environ Epidemiol, 11(3), 231–252. [DOI] [PubMed] [Google Scholar]
- Klocke C., Allen J. L., Sobolewski M., Mayer-Proschel M., Blum J. L., Lauterstein D., Zelikoff J. T., Cory-Slechta D. A. (2017). Neuropathological consequences of gestational exposure to concentrated ambient fine and ultrafine particles in the mouse. Toxicol Sci, 156(2), 492–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque S., Surace M. J., McDonald J., Block M. L. (2011. a). Air pollution and the brain: Subchronic diesel exhaust exposure causes neuroinflammation and elevates early markers of neurodegenerative disease. J Neuroinflammation, 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque S., Taetzsch T., Lull M. E., Kodavanti U., Stadler K., Wagner A., Johnson J. A., Duke L., Kodavanti P., Surace M. J., Block M. L. (2011. b). Diesel exhaust activates and primes microglia: Air pollution, neuroinflammation, and regulation of dopaminergic neurotoxicity. Environ Health Perspect, 119(8), 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. [DOI] [PubMed] [Google Scholar]
- Lieberman A., Rea W., Curtis L. (2006). Adverse health effects of indoor mold exposure. J Allergy Clin Immunol, 118(3), 763; author reply 767–768. [DOI] [PubMed] [Google Scholar]
- McCaffrey R. J., Yantz C. L. (2005). “ Cognitive impairment associated with toxigenic fungal exposure": A critique and critical analysis. Appl Neuropsychol, 12(3), 134–137. [DOI] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y., Smyth G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res, 40(10), 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell M. J., Mirer A. G., Cheung K., Tong M., Douwes J. (2011). Respiratory and allergic health effects of dampness, mold, and dampness-related agents: A review of the epidemiologic evidence. Environ Health Perspect, 119(6), 748–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Cole R. A., Gibson A. M., Bass S. A., Budelsky A. L., Reponen T., Hershey G. K. (2012). Dectin-1 and IL-17A suppress murine asthma induced by Aspergillus versicolor but not Cladosporium cladosporioides due to differences in beta-glucan surface exposure. J Immunol, 189(7), 3609–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A. P., Croston T. L., Lemons A. R., Goldsmith W. T., Marshall N. B., Kashon M. L., Germolec D. R., Beezhold D. H., Green B. J. (2018). Aspergillus fumigatus viability drives allergic responses to inhaled conidia. Ann Allergy Asthma Immunol, 121(2), 200–210.e2. [DOI] [PubMed] [Google Scholar]
- Nayak A. P., Green B. J., Lemons A. R., Marshall N. B., Goldsmith W. T., Kashon M. L., Anderson S. E., Germolec D. R., Beezhold D. H. (2016). Subchronic exposures to fungal bioaerosols promotes allergic pulmonary inflammation in naive mice. Clin Exp Allergy, 46(6), 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Gunsch C. K. (2013). Inflammatory cytokine gene expression in THP-1 cells exposed to Stachybotrys chartarum and Aspergillus versicolor. Environ Toxicol, 28(1), 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Madany A. M., Jang J. C., Valdez J. M., Rivas Z., Burr A. C., Grinberg Y. Y., Nordgren T. M., Nair M. G., Cocker D., Carson M. J., Lo D. D. (2018). Continuous inhalation exposure to fungal allergen particulates induces lung inflammation while reducing innate immune molecule expression in the brainstem. ASN Neuro, 10. 10.1177/1759091418782304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaseelan A. M., Tsilioni I., Theoharides T. C. (2018). Effects of mycotoxins on neuropsychiatric symptoms and immune processes. Clin Ther, 40(6), 903–917. [DOI] [PubMed] [Google Scholar]
- Rea W. J., Didriksen N., Simon T. R., Pan Y., Fenyves E. J., Griffiths B. (2003). Effects of toxic exposure to molds and mycotoxins in building-related illnesses. Arch Environ Health, 58(7), 399–405. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg F., Mersch-Sundermann V. (2003). Meeting report: 9th international conference on indoor air quality and climate indoor air 2002, Monterey, California, June 30–July 5, 2002. Int J Hyg Environ Health, 206(2), 133–138. [DOI] [PubMed] [Google Scholar]
- Tyler C. R., Zychowski K. E., Sanchez B. N., Rivero V., Lucas S., Herbert G., Liu J., Irshad H., McDonald J. D., Bleske B. E., Campen M. J. (2016). Surface area-dependence of gas-particle interactions influences pulmonary and neuroinflammatory outcomes. Part Fibre Toxicol, 13(1), 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper S., Wymer L. (2016). The relationship between environmental relative moldiness index values and asthma. Int J Hyg Environ Health, 219(3), 233–238. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L., Han Y., He Q. (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J Integr Biol, 16(5), 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-asn-10.1177_17590914211019886 for Aspergillus versicolor Inhalation Triggers Neuroimmune, Glial, and Neuropeptide Transcriptional Changes by Thatcher B. Ladd, James A. Johnson Jr., Christen L. Mumaw, Hendrik J. Greve, Xiaoling Xuei, Ed Simpson, Mark A. Barnes, Brett J. Green, Tara L. Croston, Chandrama Ahmed, Angela Lemons, Donald H. Beezhold and Michelle L. Block in ASN Neuro