Table 1.
Summary of 16 simulators (including our proposed scDesign2) in six properties
| Property | 1 protocol adaptive | 2 gene preserved | 3 gene cor. captured | 4 cell num. seq. dep. flexible | 5 transparent | 6 comp. and sample efficient |
|---|---|---|---|---|---|---|
| Simulator | ||||||
| dyngen [77] | ✗ | ✗ | ✓ | ✓ | ||
| Lun2 [78] | ✓ | ✗ | ✓ | ✓ | ✓ | |
| powsimR [75] | ✓ | ✓ | ✗ | ✓ | ✓ | ✓ |
| PROSST [68] | ✓ | ✗ | ✓ | ✓ | ||
| scDD [74] | ✓ | ✗ | ✗ | ✓ | ✓ | |
| scDesign[35] | ✓ | ✗ | ✓ | ✓ | ✓ | |
| scGAN [72] | ✓ | ✓ | ✗ | ✗ | ||
| splat simple[69] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ |
| splat [69] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ |
| kersplat [69] | ✓ | ✗ | ✗ | ✓ | ✓ | |
| SPARSim [71] | ✓ | ✓ | ✗ | ✓ | ✓ | |
| SPsimSeq [79] | ✓ | ✓ | ✓ |
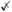 ** **
|
✓ | ✓ |
| SymSim[70] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ |
| ZINB-WaVe[39] | ✓ | ✗ | ✓ | ✓ | ||
| SERGIO [76] | ✓ | ✗* | ✓ | ✓ | ✓ | |
| scDesign2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
Property 1: protocol adaptiveness
Property 2: gene preservation
Property 3: gene correlation capture
Property 4: flexible cell number and sequencing depth choices
Property 5: transparency
Property 6: computational and sample efficiency
For each simulator and each property, a checkmark, checkcross, or cross means that the simulator satisfies, partially satisfies, or does not satisfy the property, respectively
*SERGIO requires a user-specified gene regulatory network, and it does not capture/estimate gene correlations from a real dataset
**SPsimSeq can vary cell number but not sequencing depth
