Figure 2.
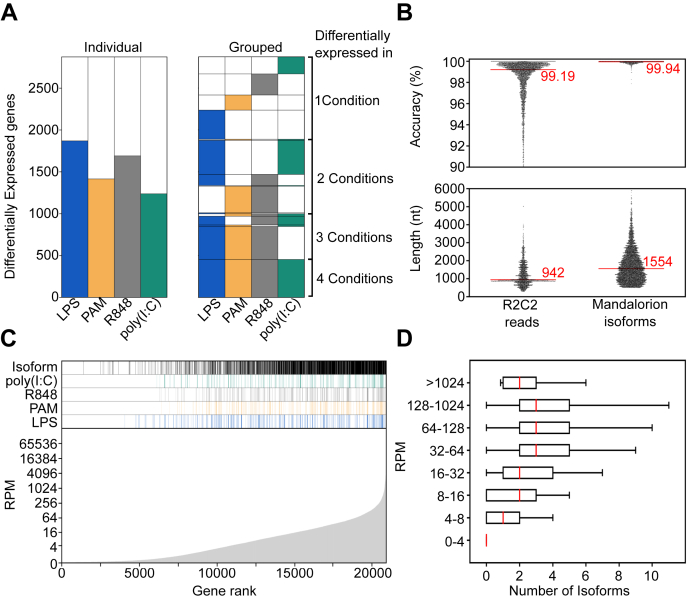
Gene and isoform level analysis.A, on the left, the numbers of genes differentially expressed between nonstimulated macrophages and macrophages stimulated with the indicated TLR ligands. On the right, genes are grouped if they were differentially expressed in more than one condition. For example, the 454 genes differentially expressed in all four conditions are shown at the bottom. Above those, the number of genes differentially expressed in LPS, PAM, and R848, but not poly(I:C) are shown. B, accuracy and length of individual R2C2 reads and Mandalorion isoforms are shown as swarmplots, with median values indicated by red lines and numeric values. C, different characteristics of genes ordered by expression are shown in this panel. From the bottom to top, this panel shows the average gene expression in reads per million (RPM) across all conditions, whether a gene is differentially expressed following LPS, Pam3CSK4 (PAM), R848, or poly(I:C) stimulation and whether we identified an isoform for the gene. D, the number of isoforms we detect for genes is shown as box plots for genes with different expression levels. TLR, toll-like receptor.