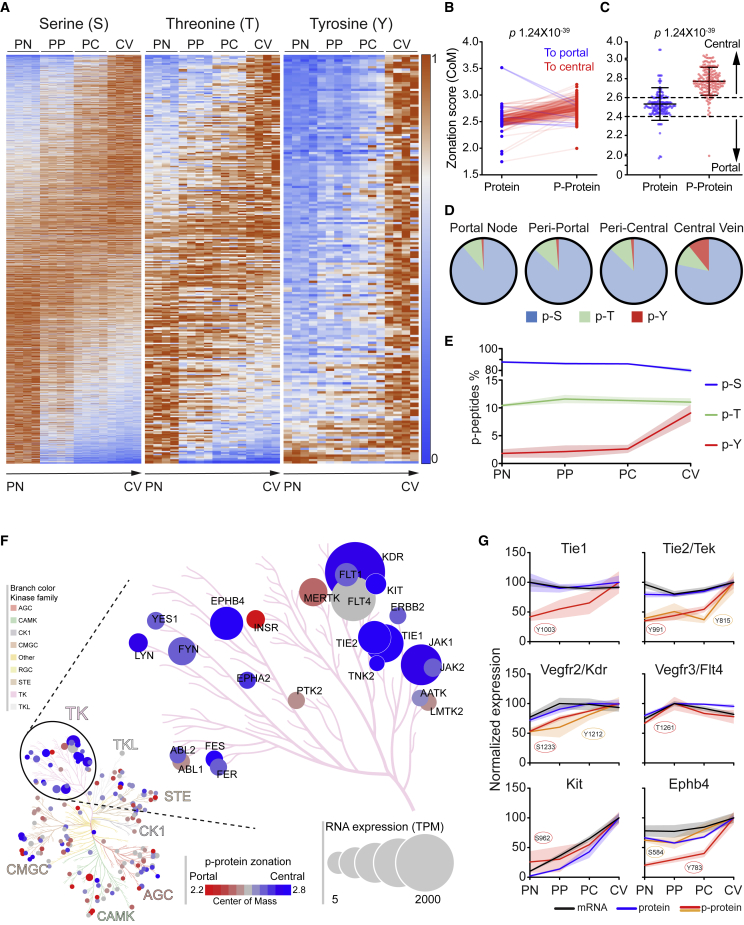
Figure 5.
Peri-central compartmentalization of tyrosine phosphorylation
(A) Heatmap representations of significantly zonated phospho-serine (P-S), phospho-threonine (P-T), and phospho-tyrosine (P-Y). P-peptides are normalized to their maximum expression and sorted by their center-of-mass (n = 4).
(B and C) Variation of the zonation score of p-Y peptides and corresponding proteins.
(B) Aligned dot plot of the CoM relative to p-Y peptides and corresponding proteins. Before-after connecting lines indicate a shift to central (red) or to portal (blue).
(C) Scatter dot plot of the same groups represented in (B). Data are represented as mean ± SD.
(D and E) Distribution of all class-I p-S, p-T, and p-Y for their strongest expressing zone. Each p-site was assigned to one zone according to the maximum expression, by average expression of four biological replicates (D) or for each replicate (E). Afterward, distribution was calculated for each zone and shown as pie charts (D) or connecting lines (E) with patches indicating SD of the four replicates.
(F) Kinome phylogenetic tree of phosphorylated kinases. Each kinase is represented by a circle and grouped by kinases family. The circle size is proportional to the corresponding TPM. The color represents phosphorylation zonation from portal (red) to central (blue).
(G) Expression profiles of matches of p-peptides (red or orange), proteins (blue), and RNA (black) for the indicated receptor tyrosine kinases. Expression is represented by percentage of maximum; patches represent SD (n = 4).
See also Figures S5 and S6.