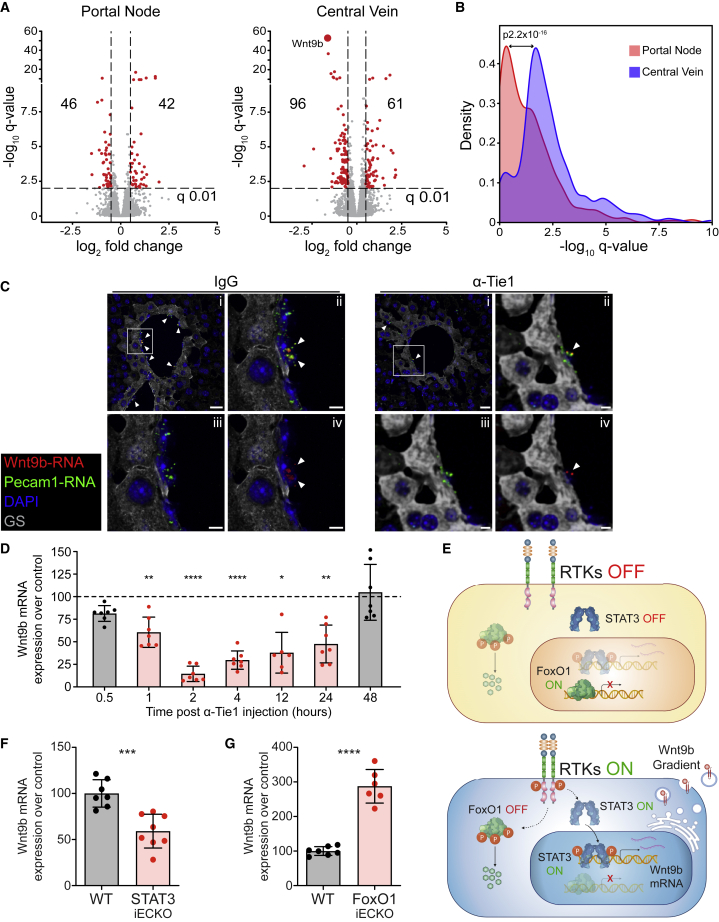
Figure 6.
CV phosphorylation of the tyrosine kinase Tie1 shapes L-EC zonation and maintains Wnt9b gradient
(A) Differential gene expression induced by Tie1 blockade. Volcano plots of gene regulations 2 h after Tie1 blockade in spatially sorted L-ECs from portal node (left) and central vein (right), respectively. Red dots mark the significantly regulated genes, indicated by the number in each square.
(B) Histogram of the −log10 q value distribution of regulated genes in portal node and/or central vein 2 h after Tie1 blockade. The effect of Tie1 blockade on PN and CV was compared by Wilcoxon matched-pairs signed rank test of the −log10 q values.
(C) RNA fluorescence in situ hybridization (FISH) analysis of Wnt9b RNA (red) 2 h after treatment of anti-Tie1 antibody, compared with IgG control. Endothelial cells were visualized by Pecam1 RNA (green) FISH staining, CV areas by glutamine synthetase (GS, gray) immunostaining, and cell nuclei (blue) counterstained with DAPI. (i) Overlay image of central vein area; (ii–iv) Zoomed overlay image (ii), Pecam1 RNA (iii), and Wnt9b RNA (iv) of the area indicated in (i). Arrow heads indicate Wnt9b RNA staining. Scale bars, 20 μm (i) and 5 μm (ii–iv).
(D) RNA expression of Wnt9b in the whole liver tissues from anti-Tie1 Ab-treated mice at the indicated time points, normalized to the relative IgG-treated mice (dashed line), significantly regulated time points highlighted in red.
(E) Signaling scheme of FoxO1 and STAT3 activation and nuclear translocation with inactive (top) or active (bottom) RTK signaling.
(F and G) RNA expression of Wnt9b in Stat3iECKO (F) and Foxo1iECKO (G) mice (red bar) normalized to the relative control mice (Cre- littermates, gray bar) from isolated L-ECs.
(C, F and G) RNA expression was determined by qRT-PCR and normalized to Actb. Data are expressed as percentage normalized to the corresponding controls. Each data point represents one animal. Data are means ± SD. Unpaired Student’s t test was used to determine the difference between experimental groups. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
See also Figure S7.