Abstract
The Coronavirus disease (COVID-19) is an infectious disease caused by the newly discovered Severe Acute Respiratory Syndrome Coronavirus two (SARS-CoV-2). Most of the people do not have the acquired immunity to fight this virus. There is no specific treatment or medicine to cure the disease. The effects of this disease appear to vary from individual to individual, right from mild cough, fever to respiratory disease. It also leads to mortality in many people. As the virus has a very rapid transmission rate, the entire world is in distress. The control and prevention of this disease has evolved as an urgent and critical issue to be addressed through technological solutions. The Healthcare industry therefore needs support from the domain of artificial intelligence (AI). AI has the inherent capability of imitating the human brain and assisting in decision-making support by automatically learning from input data. It can process huge amounts of data quickly without getting tiresome and making errors. AI technologies and tools significantly relieve the burden of healthcare professionals. In this paper, we review the critical role of AI in responding to different research challenges around the COVID-19 crisis. A sample implementation of a powerful probabilistic machine learning (ML) algorithm for assessment of risk levels of individuals is incorporated in this paper. Other pertinent application areas such as surveillance of people and hotspots, mortality prediction, diagnosis, prognostic assistance, drug repurposing and discovery of protein structure, and vaccine are presented. The paper also describes various challenges that are associated with the implementation of AI-based tools and solutions for practical use.
Keywords: Artificial intelligence, COVID-19, Deep learning, Machine learning, Applications of AI
Introduction
The new corona virus, SARS-CoV-2 which causes the disease Coronavirus disease 2019 (COVID-19) has been declared as a pandemic by WHO [1], due to its spread in almost all the countries over the world. The entire world is in crisis due to reasons including, (i) COVID-19 has rapid transmission characteristics, (ii) lack of vaccine or medicine to cure the disease, (iii) the virus easily spreads through droplets generated when an infected person coughs, sneezes, or speaks [2], (iv) An infected person can infect others even asymptomatically as well as during pre-symptomatic stage [3–5], and (v) lack of sufficient medical infrastructure, test kits, equipment, ventilators, intensive-care units (ICUs), masks, appropriate medical suits, etc., to accommodate the rising number of infected cases. Since the number of people affected is huge, performing clinical test to all the individuals is unlikely due to both availability of only limited number of testing kits [6] as well as the time involved in testing [7]. Though government authorities of many countries implement social distancing, isolation, and lockdown (curfew) to control the spread of the virus, it keeps on increasing rapidly. Due to the real situation, healthcare industry is at an urgent need to tackle the virus effectively.
AI due to its following exceptional characteristics plays a crucial role in combating various issues of COVID-19. ML algorithms are capable of learning from huge examples and extracting valuable inferences automatically in less time which facilitates faster decision-making. The availability of huge amount of historical data, advancement of hardware technologies such as multicore processors, graphical processing units (GPU), and high-speed memory helps the establishment of deeper and reinforcement learning paradigms through complex neural networks which enables object recognition, prediction, or classification with higher accuracy. Thus, AI-based tools can serve as a supplement to physicians and policy-makers to customize the healthcare plans and follow-ups according to the priorities and situation of the crisis as well as to face future pandemic situations with well-established countermeasures.
The research question of this paper is to identify the potential applications of AI to resolve the issues associated with COVID-19. Research publications have been searched via Google search engine using different keywords including ‘COVID-19’, ‘applications of AI for COVID-19’, ‘applications of machine learning and deep learning for COVID-19’, ‘digital intelligence for COVID-19’, etc. A collection of 92 publications from different electronics databases, namely, PubMed, Researchgate, arXiv, medRixv, bioRxiv, chemRxiv, and Google Scholar repositories is retrieved as in Table 1.
Table 1.
Details of publications searched via electronic databases
| S. No. | Description | Count |
|---|---|---|
| 1 | Total number of articles collected from electronic databases | 91 |
| 2 | Mathematical and statistical models for COVID-19 | 14 |
| 3 | Usefulness of the Internet of Things for COVID-19 | 6 |
| 4 | Applications of blockchain and big data for COVID-19 | 6 |
| 5 | AI for other applications | 6 |
| 6 | Number of articles relevant to the current paper | 59 |
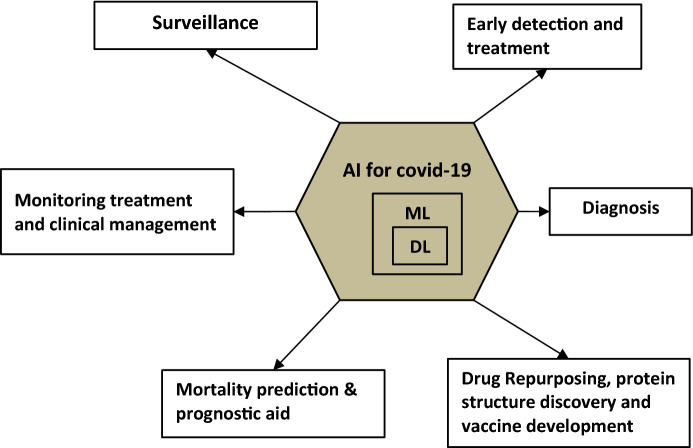
Though the publications retrieved have dealt with the theme of COVID-19 in a broader sense, the publications were found to have a wider scope to include various dimensions such as the statistical and mathematical modeling of dispersion or spread of corona virus, the applications of the internet of things (IoT) for COVID-19 issues, the applications of cloud computing and blockchain for COVID-19 challenges, the applications of microbiology, biochemistry and pathology to develop drugs and vaccines for COVID-19 along with generic survey, and AI publications. Therefore, the publications relevant to the research question are filtered out by the method of systematically reviewing the retrieved literature. That is, the collected articles are manually applied to the filtering criteria of publications that discuss the applicability of AI for combating COVID-19 issues, which resulted in 60 publications. Short descriptions of the reviewed literature are given in Table 2, along with a note on methods, strength, and weaknesses. The potential applications of AI for COVID-19 are shown in Fig. 1.
Table 2.
Summary of reviewed literature
| References | Description | Application area | Strength | Weakness |
|---|---|---|---|---|
| [8] | This work describes the applicability of AI-based tools such as Bluedot and HealthMap in detecting and giving early alert of outbreak of COVID-19 |
Surveillance Prediction of evolution pattern or trend of the pandemic Assists in implementation of preventive countermeasures |
Policy makers can implement control actions such as (i) lock down (ii) preparing testing kits (ii) identifying locations for isolating the infected case |
Difficulty in accessing data from various data sources and its integration |
| [9] | Shalow Long short-term memory based neural network is used to predict the risk category of a country into high, moderate, recovering by considering trend data and weather data combined | |||
| [10] | Prediction of COVID-19 crisis in India and comparison of it with that of Italy using machine learning approaches | |||
| [11] | AI algorithm is used to classify the data collected through mobile phone survey and classify the individuals into no, minimal, moderate and high | Early detection and assessment of COVID-19 infected cases | Cost-effective approach | Inability in detecting asymptomatic cases |
| [12] | Early prediction of COVID-19 infection using epidemiology dataset for positive and negative cases using supervised machine learning algorithms, namely, decision tree, logistic regression, Naïve Bayes, support vector machine and artificial neural network | Early prediction of COVID-19 | Prediction is performed based on epidemiology labeled dataset consisting of 2 demographic features, viz., age and sex and eight clinical features, viz., pneumonia, diabetes, asthma, hypertension, cardiovascular disease, obesity, chronic kidney disease, and tobacco | |
| [13] | RT-PCR testing is considered as the primary testing method for COVID-19 [14] and according to [13] the results produced by lung CT Scan images are highly correlated with that of RT-PCR | Diagnosis of COVID-19 |
Serves as a non-invasive testing procedure and it can identify individuals who are at high risk Accuracy of the algorithm is comparable with that of radiologist. Relieves the burden of physicians |
Needs high powerful computer hardware and huge amount of training data |
| [15, 16] | Analyzes usefulness of deep learning algorithms in medical imaging | |||
| [17] | Describes the ability of deep learning in distinguishing abnormalities of COVID-19 from other lung diseases such as pneumonia | |||
| [18, 19] | Detects the presence of COVID-19 and employs image analysis tools to map and measure the abnormalities | |||
| [20] | This work employs reinforcement learning algorithm along with deep learning in order to find optimized actions that needs to be taken according to the monitored results of treatment | Monitoring treatment | Can assist clinical decision-making such as keeping the required number of oxygen ventilated beds ready | |
| [21, 22] | The research work [21] has predicted the mortality risk due to COVID-19 using different machine learning algorithms namely, logistic regression, support vector machine, K-nearest neighbor, random forest, and gradient boosting, and found that Random Forest algorithm outperforms the other algorithms. The work [22] has identified the alarming symptoms which may lead to mortality by using different machine learning algorithms such as support vector machine (SVM), artificial neural networks, random forest, decision tree, logistic regression, and K-nearest neighbor (KNN) | Mortality prediction | To provide prognostic medical decisions | Needs to be tested over diverse datasets to make generalization |
| [23] | This work employs XGBoost supervised classifier to predict the mortality of individuals more than 10 days in advance with 90% accuracy by analyzing and determining the presence of relatively high levels of Lactic DeHydrogenase enzyme | |||
| [24] | In this work, different machine learning algorithms, namely, decision tree, random forest and support vector machine are used to predict which patients are likely to develop Acute Respiratory Distress Syndrome (ARDS), which is the severe outcome of COVID-19 from the clinical data of patients having mild COVID-19 initial presentations | |||
| [25] | According to Centers for Disease Control and Prevention, covid-19 infected individuals who are older and having chronic illnesses including heart diseases, diabetes and lung diseases are likely to have more risk. In this work, logistic regression and gradient boosted tree-based models are developed to identify the vulnerable individuals who are at severe risk | |||
| [26] | How different Deep Learning (DL) architectures help in drug discovery are discussed | Drug repurposing, Identification of protein structure and drug discovery | Significant reduction of time and cost in drug discovery | Integration of different data sources is tough |
| [27] | Deep learning-based method is proposed for drug repurposing | |||
| [28] | Machine learning models help to arrive at the knowledge about small-molecule treatment for COVID-19 by providing rapid screening of all possible therapeutic molecules which facilitates a quicker drug discovery | |||
| [29] | Deep learning algorithms are found to be efficient in finding information about the structure of protein associated with COVID-19 | |||
| [30] | Reinforcement algorithms are used to find potential candidate drugs that have the capability to inhibit the interactions of COVID-19 virus with 3C-like protease, one of the target proteins |
Fig. 1.
Potential applications of AI for COVID-19
Machine learning is a subset of AI in which statistical algorithms are trained to learn from small amount of data itself to solve a problem whereas deep learning is a subset of machine learning where deep neural network architectures are constructed to provide automatic learning of features and their representation at different hierarchical levels from huge amount of data. ML consists of different learning strategies namely supervised, unsupervised, semi-supervised, and reinforcement which are being employed according to the problem in hand. For classification and regression problems, supervised learning is employed where the algorithms are trained with labeled data to construct models. Examples for supervised algorithms include decision tree, random forest, K-nearest neighbor, support vector machine, regression, Naïve Bayes, etc. For clustering and association rule mining, unsupervised learning is used where the algorithms learn themselves from the unlabeled data according to similarity among the individual data items. In supervised learning, before training, each data item needs to be associated with its label. This consumes huge amount of time when the number of data is large. Therefore, another strategy called semi-supervised learning has been developed where at first the data are grouped into different clusters, and then, labels are annotated to the clustered in a very short time. In reinforcement learning, the algorithms are trained to take decisions in a trial-and-error method according to the environmental factors.
From the investigation made with the retrieved articles, it is found that AI provides its support for COVID-19 in different dimensions including (i) surveillance and early warnings, (ii) detection of infected individuals and assessment of risk levels, (iii) COVID-19 diagnosis, (iv) mortality prediction/prediction of individuals at high and prognostic treatment, (v) drug repurposing and drug discovery, and (vi) identification of protein structure and vaccine development. These applications are discussed in the subsequent sections. In addition, the paper discusses the challenges associated with implementation of AI-based solutions and directions for further research and development.
AI for Surveillance and Early Warnings
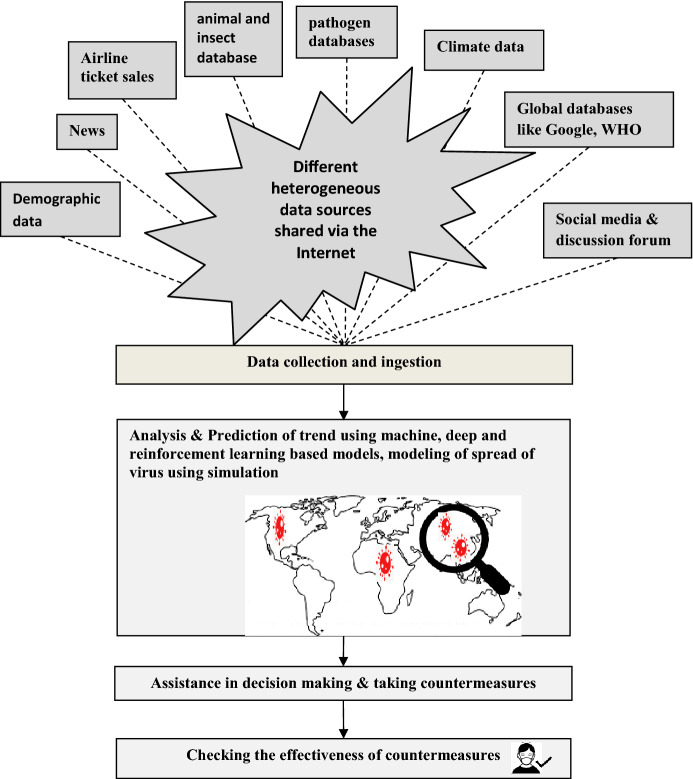
AI plays a crucial role throughout the surveillance of an epidemic, prediction of trend, providing assistance to policy-makers in taking preventive or countermeasures and assessing the effectiveness of the interventions, as shown in Fig. 2. As in Fig. 2, data are collected from different sources and analyzed using different AI techniques such as time-series and regression to identify the trend of the evolution, places which are going to be affected by the infection, when the peak of spread is likely to occur, etc.
Fig. 2.
Role of AI in surveillance
Basically, the public health surveillance is data driven and the evolving Internet and variety of open databases ease the sharing of required data to surveillance. There are several factors such as virus resistance, population density, population mobility, climatic conditions, latitude, and longitude/location which affect the spread of the virus. For example, in [32], among various parameters, namely, minimum temperature, maximum temperature, average temperature, humidity, amount of rain fall, the average temperature was found to be significantly correlated with the spread of the virus. Also, as described in [33], absolute air temperature and humidity significantly affect the spread of the virus.
Furthermore, AI-based tools and multiagent systems have the capability to mine both structured and unstructured data such as image, video, text, audio, genomic sequenced data, geographic data, data from medical devices, Electronic Health Records (EHR), and data generated by wearable devices. Also, these systems can simulate the evolution of a pandemic over time, space, and people [34, 35].
With the help of both statistical techniques such as time-series analysis and automated AI-based tools, epidemiologists along with support of data scientists are able to forecast and give daily updates on number of infected cases, number of death cases, and number of recovered cases in different countries all over the world. The potential of AI in giving early warnings can be understood with tools such as Bluedot and HealthMap [8]. The Canadian-based AI tool, BlueDot predicted the spread of Zika virus to Florida 6 months before as well as the outbreak of Ebola in West Africa. Now, in the case of COVID-19 also, this tool has identified and gave early warning by 31st December 2019 itself. This tool continuously reviews over 40 pathogen-specific datasets and integrates the data with other data resources such as news, airline ticket sales, demographic data, climate data, and animal and insect population to track over 150 infectious diseases all over the globe. In addition, another AI-based model, HealthMap at Boston Children’s hospital in USA issued an alarm on 30 December 2019 and identified the major cities that were likely to be affected by the spread. HealthMap integrates data from Google database, social media, blogs, discussion forum, etc., and predicts the epidemic.
The ability of AI-based tools in predicting the spread of a virus well in advance helps the policy-makers to plan, prioritize, and implement different interventions such as lockdown, keeping isolation centers ready, etc., according to the severity of the spread. In addition, AI also helps to check whether the public adheres to the preventive actions taken by policy-makers. For example, in Hyderabad, India, CCTV cameras which are fixed across the city are equipped with deep learning and computer vision techniques to identify the people who are not wearing face masks and will give some alerts to control center.
AI for Early Detection and Assessment
Detection of infection in affected individuals has to be done as early as possible not only for giving treatment to the infected individual but also for preventing the secondary infection. Due to lack of sufficient number of reverse transcription polymerase chain reaction (RT-PCR) testing kits and expensive cost, it becomes essential to assess the risk levels associated with individuals, so that the individuals at high risk can be given priority for testing. As mentioned in [12], phone-based surveys are analyzed using machine learning algorithms to assess the risk levels associated with individuals into different categories, namely, high risk, moderate risk, and low risk.
In this section, the usage of a machine learning algorithm to assess the risk level associated with an individual is illustrated with Naïve Bayesian classification algorithm. This algorithm is chosen as it can handle multiclass problems where more than two class labels will be involved. The problem in hand is a multiclass problem, i.e., the classifier has to predict the class label of an input data (the symptoms of COVID-19) into different risk levels or classes, namely, low, moderate, and high. In addition, this algorithm is simple and can work well even with small training data set. The algorithm assumes that the features are independent of one another. To describe the risk assessment with this algorithm, eight attributes, namely, travel (indicates an individual who had traveled abroad), contact (individual who had been in contact with people who traveled abroad), fever, cough, sneeze, shortness of breath, sore throat, and comorbidity (indicates the presence of chronic illnesses such as heart diseases, diabetes, and lung diseases) are considered [12, 36, 37].
A simple hypothetical training data set is generated as given in Table 3. For simplicity, the attributes are taken as binary attributes and can have two possible values, 0 and 1. For an attribute, the presence of 0 indicates false value and the presence of 1 indicates true value.
Table 3.
Sample training dataset
| Travel | Contact | Fever | Cough | Sneeze | Shortness of breath | Sore throat | Comorbidity | Risk level |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Low |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | Low |
| 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Moderate |
| 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Moderate |
| 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | High |
| 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | High |
| 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Moderate |
| 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | Moderate |
| 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Moderate |
| 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Moderate |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Moderate |
| 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | High |
| 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | High |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | High |
| 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | High |
| 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | Moderate |
| 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | High |
In this example, based on the attributes, the risk for COVID-19 associated with an individual is predicted using the Naïve Bayesian algorithm. Naïve Bayesian algorithm is a probability-based prediction model which works based on Bayes theorem. According to Bayes theorem, the probability of a class variable, say ‘c’ happening, given that the feature(evidence), say ‘x’ has occurred is expressed (i.e., posterior probability of class variable) as in Eq. (1)
| 1 |
where P(c|x) denotes the posterior probability of class (target) given predictor (attribute), P(c) denotes the prior probability of class, P(x|c) denotes the likelihood which is the probability of predictor, given class and P(x) denotes the prior probability of predictor.
Here, the attributes are assumed to be independent and the classification problem is framed as in (2)
| 2 |
Here, the posterior probability of a class can be calculated by first constructing a frequency table for each attribute against the target class. Then, the frequency tables are converted to likelihood tables. Finally, the posterior probability for each class is calculated using the above equation. The class with the highest posterior probability is the outcome of prediction. Please note, the denominator term, i.e., P(x) is going to be a common term while calculating the posterior probability different class labels and hence it is ignored. Therefore, the posterior probability is computed using Eq. (3)
| 3 |
Calculation of P(x|c) and P(x)
Frequency tables for the eight attributes are shown in Table 4. The likelihood tables for the above attributes (i.e., P(x|c) are given in Table 5.
Table 4.
Frequency tables for the attributes
| Frequency table for travel attribute | Frequency table for contact attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Travel | Value of the attribute | High risk | Moderate risk | No risk | Contact | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 4 | 1 | 0 | Yes | 1 | 1 | 0 | ||
| No | 3 | 7 | 2 | No | 6 | 7 | 2 | ||
| Frequency table for fever attribute | Frequency table for cough attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fever | Value of the attribute | High risk | Moderate risk | No risk | Cough | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 4 | 4 | 0 | Yes | 2 | 2 | 0 | ||
| no | 3 | 4 | 2 | No | 5 | 6 | 2 | ||
| Frequency table for sneeze attribute | Frequency table for shortness of breath attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sneeze | Value of the attribute | High risk | Moderate risk | No risk | Shortness of breath | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 2 | 1 | 0 | Yes | 1 | 1 | 0 | ||
| no | 5 | 7 | 2 | No | 6 | 7 | 2 | ||
| Frequency table for sore throat attribute | Frequency table for comorbidity attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sore throat | Value of the attribute | High risk | Moderate risk | No risk | Comorbidity | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 1 | 2 | 0 | Yes | 4 | 2 | 1 | ||
| No | 6 | 6 | 2 | No | 3 | 6 | 1 | ||
Table 5.
Likelihood tables for attributes (i.e., P(x|c))
| Likelihood table for Travel attribute | Likelihood table for contact attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Travel | Value of the attribute | High risk | Moderate risk | No risk | Contact | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 4/7 | 1/8 | 0/2 | Yes | 1/7 | 1/8 | 0/2 | ||
| No | 3/7 | 7/8 | 2/2 | No | 6/7 | 7/8 | 2/2 | ||
| Likelihood table for fever attribute | Likelihood table for cough attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fever | Value of the attribute | High risk | Moderate risk | No risk | Cough | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 4/7 | 4/8 | 0/2 | Yes | 2/7 | 2/8 | 0/2 | ||
| no | 3/7 | 4/8 | 2/2 | No | 5/7 | 6/8 | 2/2 | ||
| Likelihood table for sneeze attribute | Likelihood table for shortness of breath attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sneeze | Value of the attribute | High risk | Moderate risk | No risk | Shortness of breath | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 2/7 | 1/8 | 0/2 | Yes | 1/7 | 1/8 | 0/2 | ||
| no | 5/7 | 7/8 | 2/2 | No | 6/7 | 7/8 | 2/2 | ||
| Likelihood table for sore throat attribute | Likelihood table for comorbidity attribute | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sore throat | Value of the attribute | High risk | Moderate risk | No risk | Comorbidity | Value of the attribute | High risk | Moderate risk | No risk |
| Yes | 1/7 | 2/8 | 0/2 | Yes | 4/7 | 2/8 | 1/2 | ||
| No | 6/7 | 6/8 | 2/2 | No | 3/7 | 6/8 | 1/2 | ||
Calculation of P(c)
In Table 3, the column, risk level refers to target class label and it can have three values, high, moderate, and low. Prior probability of class [i.e., P(c)] is computed and given in Table 6.
Table 6.
Prior probability of class, P(c)
| Risk level | P(c) |
|---|---|
| High | P(high) = 7/17 |
| Moderate | P(moderate) = 8/17 |
| Low | P(low) = 2/17 |
Calculation of Posterior Probability, P(c|x)
Now, consider test dataset as given in Table 7.
Table 7.
Test dataset
| Travel | Contact | Fever | Cough | Sneeze | Shortness of breath | Sore throat | Comorbidity | Actual risk level |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | Moderate |
| 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Moderate |
| 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Moderate |
Now, consider the first record as X. To find the class label for X which is having attribute values, travel = no, contact = no, fever = yes, cough = yes, sneeze = yes, shortness of breath = no, sore throat = no, comorbidity = no, posterior probabilities for different risk levels are calculated as given through Eqs. (4), (5) and (6). Then, the class which has the highest value for probability is predicted as class of the given record
| 4 |
| 5 |
Similarly
| 6 |
| 7 |
Now, the posterior probabilities are normalized between 0 and 1 as given in Eqs. (8), (9) and (10)
| 8 |
| 9 |
| 10 |
Since, the moderate class is having highest probability, for the given record, X, the class is predicted as ‘moderate’.
Similarly, the class label has been predicted for the remaining two records as ‘moderate’ and ‘moderate’, respectively.
The accuracy of the above hypothetical example is 100% as the predicted classes exactly match with those of actual class labels.
In real application, the input dataset is split into training data and test data where training data are used to train the algorithm. The training data should include that are likely to occur. The training dataset should be carefully chosen to all possible situations and cases as the quality of training dataset is important in determining the accuracy of the algorithm. After training, the algorithm needs to be employed over test dataset to verify the adequacy of accuracy of the algorithm for a given problem. Along with accuracy, precision, recall, and f_score are also used to validate the performance of the model. The first step of evaluation is to construct the confusion matrix. The second step is to compute different measures from the confusion matrix. The computation of different evaluation measures for a multiclass problem is shown with sample of 100 test records with assumed values for different classes as in Table 8.
Table 8.
Confusion matrix for a multiclass problem with three classes high, moderate, and low
| Actual high | Actual moderate | Actual low | Total predicted | |
|---|---|---|---|---|
| Predicted high | 80 | 10 | 0 | 90 |
| Predicted moderate | 10 | 90 | 0 | 100 |
| Predicted low | 10 | 0 | 90 | 100 |
| Total actual | 100 | 100 | 90 | 290 |
After constructing the confusion matrix, the values for precision and recall can be calculated using two methods, microaverage and macroaverage [16]. In microaverage method, individual true positive, true negative, false positive, and false negate are added up for all class labels and then average is taken. In macroaverage method, precision and recall values for each class are computed, and then, the average is taken. To provide an insight to the reader, the macroaverage method is discussed below.
With values given in Table 8, the precision and recall for the individual classes can be computed.
Now, consider the class label, high. Recall and precision for this class are computed as
Similarly, recall and precision for the other classes moderate and low are computed as follows:
Now, the precision and recall for the models are computed by taking the average of the individual class values. Therefore, precision of the model is average_of (88.8 + 90 + 90) = 89.6%. Similarly, the recall of the model is average_of (80 + 90 + 100) = 90%.
Another interesting point about Naïve Bayesian algorithm is that the value of probability gives the confidence of prediction also. Thus, machine learning algorithms can be used to assess risk level associated with individuals. With the knowledge about risk levels of individuals, individuals at high risk can be given higher priority for testing as well as hospital-based medical treatment. Furthermore, this kind of preliminary assessment using machine learning algorithms serves as a non-invasive and non-contact method of screening, which too reduces the spread of infection. It helps in keeping ready the required number of medical equipment for the needy.
AI for Diagnosis
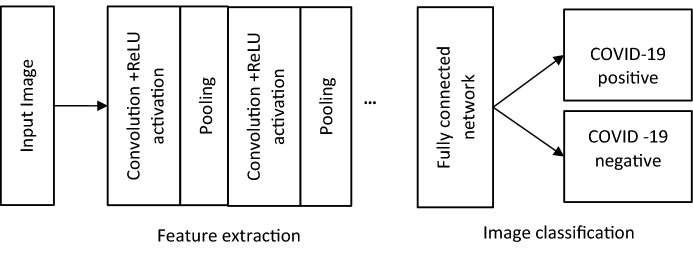
As mentioned earlier, lung CT scan is one of the testing procedures for detecting COVID-19 infection in lungs. Convolutional Neural Network (CNN), a kind of neural network which takes images as input, assigns importance to various aspects of the image and when taught with huge number of examples with annotated labels, can classify the images. The typical design of CNN for image classification is shown in Fig. 3 [38]. The design of input and output layers is simple and straight forward. The input layer would contain the values of pixels of lung CT scan images. For example, if the image is pixels, then there will be 4096 inputs having values of intensity ranging from 0 to 1.
Fig. 3.
Higher level CNN-based deep learning for COVID-19 classification
In CNN, one or more convolutional and pooling layers are used to perform extraction of features such as edges, lines, etc., and reduction of spatial representation of images, respectively [15]. Basically, the convolutional layer applies a filter that moves over the input image and extracts features from the input image. Filters use or kernels which when moves over the input image, performs element-wise multiplication and addition. The number of pixels needs to be shifted is determined by a parameter called stride. There are different types of filters which extract different features, namely edges, lines, etc. The convolutional layer includes a rectified linear unit (ReLU) activation function which is used to bring non-linear transformation in the network. Pooling layer is responsible for reducing the spatial size of the convolved feature to reduce the computational power required to process the data through dimensionality reduction. There are different types of pooling, namely, Max Pooling, which returns the maximum value from the portion of the image covered by the filter and Average Pooling which returns the average of all the values from the portion of the image covered by the filter. Fully connected serves as a classifier. It uses extracted features and predicts the label of the image.
An important feature of CNN is that weights of these filters are automatically learnt and fixed during training. All these extracted features then are ‘combined’ to form feature maps which enable the image classification in the fully connected layer. Typically, in healthcare applications, to ensure high accuracy, deep learning algorithms are designed in supervised learning model. That is, the input images are annotated with corresponding class labels and given for learning. When deep learning algorithm is given a huge number of training images with annotations, the algorithm will learn through the hidden, connected neural network and will try to detect the common patterns/similarities from the images with similar labels. Once the learning step is complete, then the algorithm can predict the type of unknown image and classify the label of unknown image. In addition to image classification, as in [18], in the case of COVID-19-positive images, the abnormalities and ground glass opacities (GGO) are mapped and measured using 2D slice analysis and 3D volume analysis tools which are then visualized with 3D visualization tools. Ultimately, deep learning can analyze a huge number of CT scans per day and reduce the burden lying with the radiologist; also, the performance of the algorithm is comparable to an expert radiologist as in [19, 31].
AI for Mortality Prediction and Prognostic Support
Predicting mortality risk is very important in the case of COVID-19, as the number of infected cases goes on increasing very rapidly. Obviously, both developed and developing countries do not have sufficient medical infrastructure to accommodate all the affected individuals. In such situation, it becomes mandatory to have a knowledge on (i) how many individuals are at high risk, (ii) number of individuals who are already having chronic diseases such as heart disease, diabetes, and lung diseases, (iii) number of individuals who are in need of ventilators, and (iv) number of individuals who are in need of ICUs, etc.
As mentioned earlier, research works [21–26] have employed AI algorithms to predict people who are at mortality risk and provide information about the same well in advance. For example, as in [21], an open-source tool using Random Forest algorithm has been deployed online through https://ashis-das.shinyapps.io/CoCoMoRP/ for mortality prediction which enables appropriate planning for keeping the required number of ventilators ready. In [22], neural network-based model has been developed using data collected from different hospitals, all around the world, which predicts mortality with an accuracy of 89.98%, based on demographic information, physiological data, patient’s symptoms, and pre-existing conditions. Three features, namely, LDH, hs-CRP, and lymphocytes, have been identified as predictors for mortality based on clinical data collected from China [23]. Also, XGBoost algorithm was found to predict mortality rate more than 10 days in advance and then assists in prognostic treatment. In [39], machine learning techniques have been applied to large collection of patient-level data to predict mortality based on three clinical features, namely, patient’s age, minimum oxygen saturation, and type of patient, i.e., inpatient or outpatient. Experimentation with two different datasets of the work exhibits that the patients who died are older with mean age of 73.4 years and had lower oxygen saturation at their presentation to hospitals. Also, they had comparatively later encounter at a hospital. Patients who died are likely to have hypertension and diabetes. Alternate to mortality prediction, in the research work [40], different data mining models such as decision tree, support vector machine, naïve Bayes, etc., have been used to predict the minimum and maximum number of days for recovery of the patients with respect to age of the patients. This study predicted that the patients of age from 65 to 85 are at high risk of recovery from the disease.
AI for Repurposing Drugs
Repurposing drugs is a method of using existing drugs for the treatment of other new diseases for which the drugs had not been approved. It provides a very cost-effective and less time-consuming approach for identifying existing drugs for emerging diseases. As seen from Fig. 4 and Fig. 5, the timeline (excluding Food and Drug Administration post-market safety monitoring) involved in the conventional drug discovery is very long of around 15 years where it is only around 3 to 10 years in drug repurposing [41]. The time spent in initial drug discovery and pre-clinical trial stages are greatly reduced in drug repurposing [42, 43]. In addition, risks and cost involved in drug repurposing also greatly reduced.
Fig. 4.
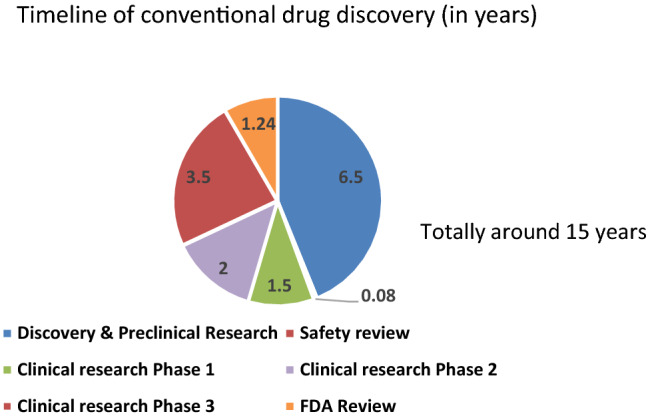
Timeline in conventional drug discovery
Fig. 5.
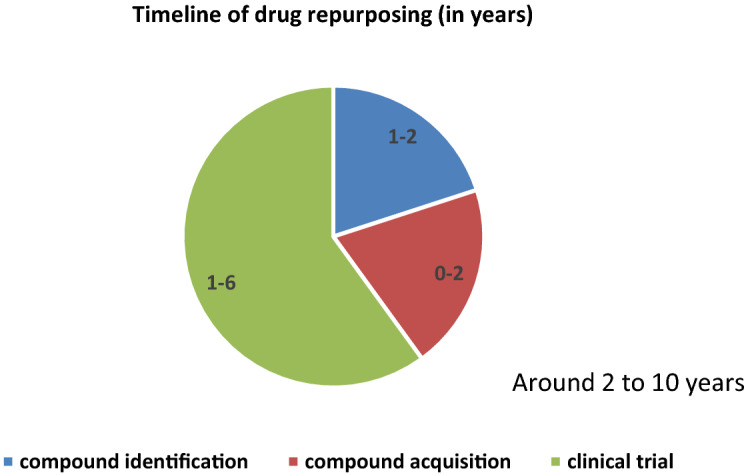
Time line in drug repurposing
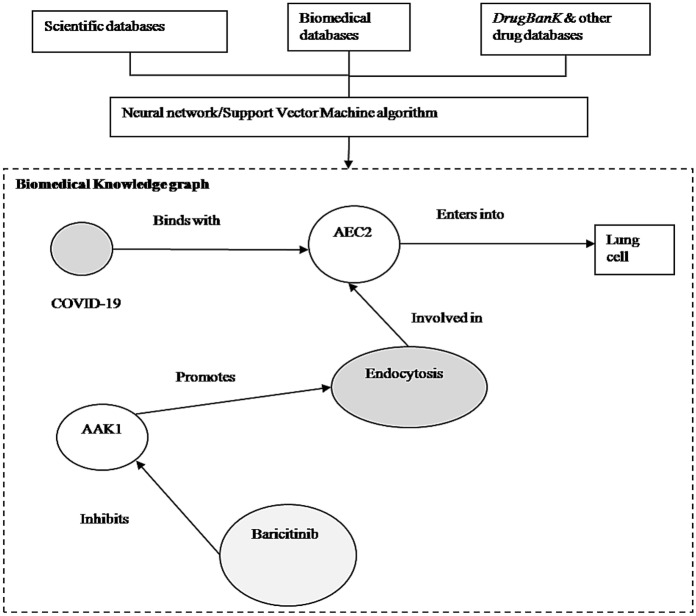
In compound identification stage, computational approaches are efficient in identifying the exiting drugs. AI-based models are used to scan through drug and disease databases containing terabytes of published and unpublished data to construct biomedical knowledge graph with more than 31 million biomedical disease and drug concepts. The graph provides clues in identifying the existing drugs which inhibit the viral infection into lungs. Consider the portion of the knowledge graph, as shown in Fig. 6. It is understood that that the virus binds to a particular protein called ACE2 to enter into lung cells. This biological process is called endocytosis which is regulated by another protein called AAK1. As in Fig. 6, it is clear that the biomedical graph helps in identifying a number of compounds that inhibit AAK1, including Baricitinib which is already licensed to treat rheumatoid arthritis. In addition to knowledge graph-based approach, there are various other approaches namely, molecular docking, target, pathway mapping, machine learning, artificial neural network, deep learning and text mining-based approaches as discussed in [44]. The computational methods are very effective in the high level integration of existing knowledge and data which in turn identifies existing drugs for treatment of new diseases along with side effects and other interactions. In the case of COVID-19, the computational methods help in identifying potential drugs including chloroquine, remdesivir, ritonavir, etc. [43].
Fig. 6.
A portion of biomedical knowledge graph
Furthermore, AI also helps in extracting information about the adverse effects that may exist when two or more drugs are taken together with the help of entity linking tools such as Falcon. For example, this tool enables the understanding of the potential adverse medical conditions that may occur when an existing drug is tried in the presence of chronic conditions such as high blood pressure, asthma, or diabetes. Furthermore, AI models are trained to explore a huge chemical space and they automatically screen the huge space and identify the most potential candidates based on different parameters like opportunity, expected response reliability, and safety. For example, as discussed in [30], a reinforcement learning approach is used to identify potential candidates to inhibit the interactions of the virus with 3C-like protease, one of the target proteins for which the crystal structure is known. The algorithm helped in identifying 284 molecules to inhibit the interaction which will further be explored for development of new drugs.
AI for Study of Protein Structure and Vaccine Development
In general, vaccines imitate the virus to produce defensive white blood cells in our body. Vaccines are of different types, namely, whole pathogen vaccine designed using killed or weakened pathogens, subunit vaccine which use part of the germ like protein, and nucleic acid vaccine which contain the genetic material of the germ. Here, AI is useful in accelerating the development of subunit and nucleic acid vaccines. Determining the structure of a protein is essential in understanding the function of a protein which in turn facilitates the development of vaccines. However, a single protein is composed of several amino acids and determining the structure of a protein is tedious. When an antigen (say, COVID-19) enters a human body, it causes the immune system of a body to generate antibodies, specialized proteins which fight against antigen. Different kinds of antibodies including IgG, IgA, IgM, IgE, and IgD will get generated according to the antigen entered into a human body. Each antibody has a unique binding site shape which locks onto the specific shape of the antigen. Then, the antibodies destroy the antigen. An epitope is a cluster of amino acids found on the outside of the antigen and is the part of an antigen that is recognized by the antibodies. Finding and classifying epitopes are essential in developing new drugs.
Machine learning algorithms such as Support vector machines (SVM), hidden Markov Models, and artificial neural networks (specifically deep learning) have all proven to be faster and more accurate at identifying epitopes [45]. For example, Google DeepMind introduced Alphafold system to find protein structure of COVID-19 virus with Deep Residual Network. Furthermore, the AI models are helpful in analyzing the potential mutations of the virus and corresponding identifying the best possible candidates for vaccine design [46].
Challenges and Conclusion
Despite the capabilities of AI models in finding combating solutions for COVID-19, it is very difficult to get all kinds of relevant data that are required to build such models. For example, research works such as [21–25] need real patient-level data from different hospitals data (after depersonalization) while building the models, but it is not easy to access the hospital data due to Health Insurance Portability and Accountability Act of 1996 (HIPAA) restrictions. It becomes challenging to provide relevant and right data to the models. Furthermore, algorithms such as deep learning should be trained with large, accurately labeled, curated datasets to produce sufficient accuracy. As described in [36], choosing the right dataset from various open online databases of chest CT and chest X-ray datasets becomes more challenging and as a key to the above challenge, transfer learning is being used while building models for COVID-19.
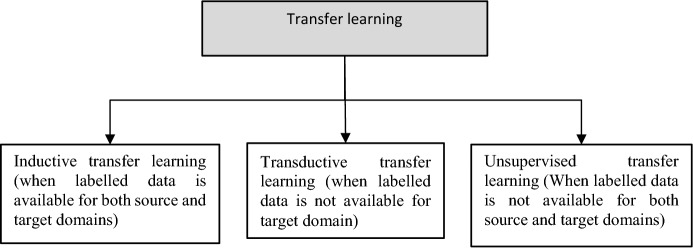
In conventional machine learning methods, the models are isolated which means that there is no transfer of knowledge from one model to another model. However, in reality, humans always tend to use the past learnt knowledge while solving new tasks. For example, when one learns the driving of four wheelers, he uses his previous of knowledge of driving two wheelers (and he does not start from the scratch). In transfer learning, the model developed for a task is reused for solving another task. This kind of transfer learning can be used whenever the available data are small as well as when the training time is longer. Here, the detection of COVID-19 can be enhanced using transfer learning. According to the similarity between the domains and availability of data for source and target problems involved in transfer learning, different learning strategies, namely inductive, transductive, and unsupervised, as shown in Fig. 7 are being used.
Fig. 7.
Different techniques of transfer learning
Also, data augmentation operations, such as flip, rotation, scale, crop, translation, etc., are also being used to expand the data size and thereby reduce the overfitting issues and help in enhancing the performance of the models [47]. In addition, when training data are constructed from online resources, the data are likely to contain false data also which will bias the accuracy of AI-based models. Furthermore, data retrieved from different sources may conflict with one another which poses additional challenge in arriving at correct information to train the algorithms [48].
Existing deep learning models lack the explanation for their detection or classification of inputs, so the physicians may not be able to understand why the models have predicted as they did [49, 50]. Therefore, the black box nature of the models needs to be improved with explainable AI. A deep learning approach presented in [51] is explainable by its design. The approach provides reasons for why it predicts the label for a given data as it does, using a set of IF–THEN rules. In addition to the explainability or interpretability, the proposed approach has the ability to actively learn from new data samples.
Despite the challenge of implementation of transfer learning and explainability in the models, another big challenge exists with respect to the validation of the developed models. Most of the research works use a specific dataset for the proof of concept. Radiologists elsewhere have expressed that most of COVID-19 images come from Chinese hospitals, and hence, the developed models tend to suffer from selection bias [52]. Therefore, before put into practical use, the developed models are required to be tested over diverse data sets which cover a wider demographic feature, so that the models can be generalized and tools can be developed [53].
Despite AI is an enabling technology to support diagnosis and prognosis, it is dependent on various factors such as appropriate infrastructure, authenticated and trustable data sharing along with unbiased safe translation of models into usable solutions. Finding combating solutions for COVID-19 needs combined efforts from international cross-disciplinary collaborations to carefully identify time, course, and region-dependent clinical actions in response to COVID-19 [54]. Furthermore, the AI needs to go in hand with other digital intelligence technologies such as the wearable technology, mobile technology, the Internet of Things, big data analytics, cloud computing [55–59] to develop viable, thoroughly validated tools which can provide real support local healthcare providers. In addition, as discussed in the paper, AI has its applications in finding solutions for COVID-19 in many dimensions.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chellammal Surianarayanan, Email: chelsganesh@gmail.com.
Pethuru Raj Chelliah, Email: peterindia@gmail.com.
References
- 1.The Lancet Infectious Diseases COVID-19, a pandemic or not? Lancet Infect. Dis. 2020;20(4):383–383. doi: 10.1016/S1473-3099(20)30180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease 2019 (COVID-19) Situation Report—40, WHO, 29 Feb 2020. [Online] https://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/update-20-epi-win-covid-19.pdf?sfvrsn=5e0b2d74_2
- 3.He X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 4.Tong Z, et al. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg. Infect. Dis. 2020;26(5):1052–1054. doi: 10.3201/eid2605.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi, M et al: Asymptomatic transmission, the Achilles’ heel of current strategies to control COVID-19. N. Engl. J. Med. [Online] 10.1056/NEJMe2009758, Available: https://www.nejm.org/doi/full/10.1056/NEJMe2009758 [DOI] [PMC free article] [PubMed]
- 6.Raza, K.: Artificial intelligence against COVID-19: a meta-analysis of current research. In: Hassanien, A.E., Dey, N., Elghamrawy, S. (eds.) Big data analytics and artificial intelligence against COVID-19: innovation vision and approach, studies in big data, vol 78. Springer, Cham 2020 10.1007/978-3-030-55258-9_10
- 7.Green, K. et al: CEBM, Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. 7 April 2020, [Online]: https://www.cebm.net/Covid-19/molecular-and-antibody-point-of-care-tests-to-support-the-screening-diagnosis-and-monitoring-of-Covid-19/
- 8.Long JB, Ehrenfeld JM. The role of augmented intelligence (AI) in detecting and preventing the spread of novel Coronavirus. J Med Syst. 2020;44:59. doi: 10.1007/s10916-020-1536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pal, R., Sekh, A.A., Kar, S., Prasad, D.K.: Neural network based country wise risk prediction of COVID-19. IEEE Access, arXiv: 2004.00959v1 [q-bio.PE] (2020)
- 10.Tiwari UK, Khan R. Role of machine learning to predict the outbreak of COVID-19 in India. Xi'an Jianzhu Keji Daxue Xuebao/J. Xi'an Univ. Archit. Technol. 2020;12(4):2663–2669. [Google Scholar]
- 11.Srinivasa Rao ASR, Vazquez JA. Identification of COVID-19 can be quicker through artificial intelligence framework using a mobile phone–based survey when cities and towns are under quarantine. Infect. Control Hosp. Epidemiol. 2020 doi: 10.1017/ice.2020.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muhammad LJ, Algehyne EA, Usman SS, et al. Supervised machine learning models for prediction of COVID-19 infection using epidemiology dataset. SN Comput. Sci. 2021;2:11. doi: 10.1007/s42979-020-00394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aiet T, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Udugama B, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 15.Rawat W, Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Commun. 2017;29:2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 16.Lee J-G, Jun S, Cho Y-W, Lee H, Kim GB, Seo JB, Kim N. Deep learning in medical imaging: general overview. Korean J. Radiol. 2017;18(4):570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Qin L, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Thorac. Imaging. 2020 doi: 10.1148/radiol.2020200905. [DOI] [Google Scholar]
- 18.Gozes, O. et al.: Rapid AI development cycle for the Coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. Radiol. Artif. Intell. https://arxiv.org/ftp/arxiv/papers/2003/2003.05037.pdf
- 19.Wang S, et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) Eur. Radiol. 2021 doi: 10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dell’Aversana, P.: Deep neural networks and reinforcement learning for studying COVID-19 pandemic. 2020, Researchgate: https://www.researchgate.net/publication/339953916_Deep_Neural_Networks_and_Reinforcement_Learning_for_studying_COVID-19_pandemic
- 21.Das A, Mishra S, Gopalan SS. Predicting community mortality risk due to CoVID-19 using machine learning and development of a prediction tool. medRxiv. 2020 doi: 10.1101/2020.04.27.20081794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pourhomayoun M, Shakibi M. Predicting mortality risk in patients with COVID-19 using artificial intelligence to help medical decision-making. medRxiv. 2020 doi: 10.1101/2020.03.30.20047308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L, Zhang H, Goncalves J, et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2:283–288. doi: 10.1038/s42256-020-0180-7. [DOI] [Google Scholar]
- 24.Jiang X, Coffee M, Bari A, Wang J, Jiang X, et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. CMC Comp. Mater. Continua. 2020;63(1):537–551. [Google Scholar]
- 25.De Capprio D, Gartner J, McCall CJ, Burgess T, Kothari S, Sayed S. Building a COVID-19 vulnerability index. medrxiv Repos. 2020 doi: 10.1101/2020.03.16.20036723. [DOI] [Google Scholar]
- 26.Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov. Today. 2018;23(6):1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 27.Zhavoronkov A, et al. Potential COVID-2019 3C-like protease inhibitors designed using generative deep learning approaches. ChemRxiv Preprint. 2020 doi: 10.26434/chemrxiv.11829102.v2. [DOI] [Google Scholar]
- 28.Batra, R et al: Screening of therapeutic agents for COVID-19 using machine learning and ensemble docking simulations. Researchgate Repos. (2020) [DOI] [PubMed]
- 29.Senior AW, et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 30.Tang, B., He, F., Liu, D., Fang, M., Wu, Z., Xu, D.: AI aided design of novel targeted covalent inhibitors against SARS-CoV-2. bioRxiv preprintbioRxiv:2020.03.03.972133 (2020) [DOI] [PMC free article] [PubMed]
- 31.Jin S, Wang B, Haibo X, Luo C, Wei L, Zhao W, Hou X, Ma W, Zhengqing X, Zheng Z, Sun W, Lan L, Zhang W, Xiangdong M, Shi C, Wang Z, Lee J, Jin Z, Lin M, Jin H, Zhang L, Guo J, Zhao B, Ren Z, Wang S, You Z, Dong J, Wang X, Wang J, Wei X. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. medRxiv. 2020 doi: 10.1101/2020.03.19.20039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosepua R, Gunawan J, Effendy DS, Ahmad LOAI, Lestari H, Bahar H, Asfian P. Correlation between weather and COVID-19 pandemic in Jakarta Indonesia. Sci. Total Environ. 2020;725:138436. doi: 10.1016/j.scitotenv.2020.138436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Liang H, Yuan X, Hu Y, Xu M, Zhao Y, et al. Roles of meteorological conditions in COVID-19 transmission on a worldwide scale. medRxiv. 2020;382:727. [Google Scholar]
- 34.https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-criteria.html?CDC_AA_refVal%3dhttps%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fhcp%2Fidentify-assess-flowchart.html
- 35.Zeng D, Cao Z, Neill DB. Artificial intelligence–enabled public health surveillance—from local detection to global epidemic monitoring and control. Artif. Intell. Med. 2021 doi: 10.1016/B978-0-12-821259-2.00022-3. [DOI] [Google Scholar]
- 36.Raza, K., Maryam, Qazi S.: An introduction to computational intelligence in COVID-19: surveillance, prevention, prediction, and diagnosis. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. studies in computational intelligence, vol 923. Springer, Singapore. (2021) 10.1007/978-981-15-8534-0_1
- 37.Asif HM, Hashmi HAS. Early detection of COVID-19. medrxiv Repos. Paper. 2020 doi: 10.1101/2020.04.04.20053363. [DOI] [Google Scholar]
- 38.Mansour NA, Saleh AI, Badawy M, et al. Accurate detection of COVID-19 patients based on feature correlated Naïve Bayes (FCNB) classification strategy. J. Ambient. Intell. Human. Comput. 2021 doi: 10.1007/s12652-020-02883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadaw AS, Li YC, Bose S, Iyengar R, Bunyavanich S, Pandey G. Clinical features of COVID-19 mortality: development and validation of a clinical prediction model. Lancet Digit. Health. 2020;2(10):e516–e525. doi: 10.1016/S2589-7500(20)30217-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam LJ, Usman MM, Usman SS, et al. Predictive data mining models for novel coronavirus (COVID-19) infected patients' recovery. SN Comput. Sci. 2020;1:206. doi: 10.1007/s42979-020-00216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue H, Li J, Xie H, Wang Y. Review of drug repositioning approaches and resources. Int. J. Biol. Sci. 2018;14(10):1232–1244. doi: 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohanty S, Harun-Ai-Rashid M, Mridul M, Mohanty C, Swayamsiddha S. Application of artificial intelligence in COVID-19 drug repurposing. Diabetes Metab. Syndr. 2020;14(5):1027–1031. doi: 10.1016/j.dsx.2020.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahu, A., Qazi, S., Raza, K., Verma, S.: COVID-19: hard road to find integrated computational drug and repurposing pipeline. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. studies in computational intelligence, vol 923, (2021), Springer, Singapore. 10.1007/978-981-15-8534-0_15
- 44.Tripathi, M.K., Sharma, S., Singh, T.P., Ethayathulla, A.S., Kaur, P.: Computational intelligence in drug repurposing for COVID-19. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. studies in computational intelligence, vol 923. Springer, Singapore (2021) 10.1007/978-981-15-8534-0_14
- 45.Bharadwaj, K.K. et al: Computational intelligence in vaccine design against COVID-19. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: Surveillance, prevention, prediction and diagnosis. studies in computational intelligence, vol 923. Springer, Singapore. (2021) 10.1007/978-981-15-8534-0_16
- 46.Arora N, Banerjee AK, Narasu ML. The role of artificial intelligence in tackling COVID-19. Future Virol. 2020 doi: 10.2217/fvl-2020-0130. [DOI] [Google Scholar]
- 47.Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S, Shukla PK. Deep transfer learning based classification model for COVID-19. IRBM. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alafif T, Tehame AM, Bajaba S, Barnawi A, Zia S. Machine and deep learning towards COVID-19 diagnosis and treatment: survey, challenges, and future directions. Int. J. Environ. Res. Public Health. 2021;18(3):1117. doi: 10.3390/ijerph18031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh RK, Pandey R, Babu RN. COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput. Appl. 2021 doi: 10.1007/s00521-020-05636-6]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.medRxiv preprint doi: 10.1101/2020.04.24.20078584
- 52.Wim N. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. AI & Soc. 2020 doi: 10.1007/s00146-020-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kallianos K, Mongan J, Antani S, Henry T, Taylor A, Abuya J, et al. How far have we come? Artificial intelligence for chest radiograph interpretation. Clin. Radiol. 2019;74(5):338–345. doi: 10.1016/j.crad.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y, Jacob J, Parker GJM, et al. The challenges of deploying artificial intelligence models in a rapidly evolving pandemic. Nat. Mach. Intell. 2020;2:298–300. doi: 10.1038/s42256-020-0185-2. [DOI] [Google Scholar]
- 55.Islam MM, Mahmud S, Muhammad LJ, Islam MR, Nooruddin S, Ayon SI. Wearable technology to assist the patients infected with novel coronavirus (COVID-19) SN Comput. Sci. 2020;1(6):320. doi: 10.1007/s42979-020-00335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmad, S. et al: Mobile technology solution for COVID-19: surveillance and prevention. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. Studies in computational intelligence, vol 923. Springer, Singapore (2021), 10.1007/978-981-15-8534-0_5
- 57.Mohammed, I.B., Isa, S.M.: The role of internet of things (IoT) in the containment and spread of the novel COVID-19 pandemic. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. Studies in computational intelligence, vol 923. Springer, Singapore (2021) 10.1007/978-981-15-8534-0_6
- 58.Aggarwal, S., Singh, N., Mishra, K.K.: Digital image analysis is a silver bullet to COVID-19 pandemic. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. Studies in computational intelligence, vol 923. Springer, Singapore, (2021) 10.1007/978-981-15-8534-0_21
- 59.Verma, S., Gazara, R.K.: Big data analytics for understanding and fighting COVID-19. In: Raza, K. (eds.) Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis. Studies in computational intelligence, vol 923. Springer, Singapore, (2021), 10.1007/978-981-15-8534-0_17