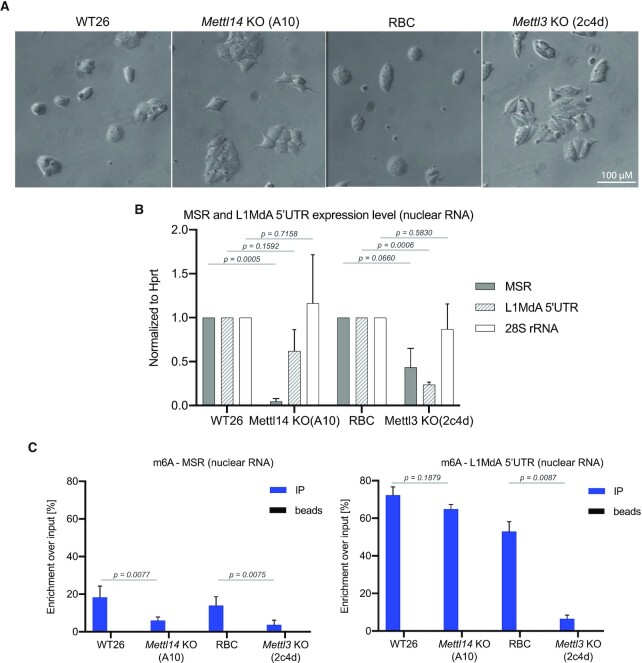
Figure 3.
Mettl14 and Mettl3 mutant ES cells display reduced levels of MSR m6A RNA methylation. (A) Morphology of WT26, Mettl14 KO (clone A10), RBC and Mettl3 KO (clone 2c4d) ES cells under bright-field microscopy. Scale bar = 100 μM. (B) Histogram showing the expression level of MSR (grey bar), LINE L1MdA 5′UTR (hatched bar) and 28S rRNA (white bar) transcripts in WT26, Mettl14 KO (clone A10), RBC and Mettl3 KO (clone 2c4d) ES cells. Expression was analyzed in nuclear RNA by RT-qPCR and normalized to the expression of Hprt in WT26 cells. The data represent the mean ± SD from n = 3 biological replicates. (C) MeRIP analysis of m6A-enriched MSR (left) and LINE L1MdA 5′UTR (right) transcripts in nuclear RNA from WT26, Mettl14 KO, RBC and Mettl3 KO ES cells. Enrichment is calculated as the percentage of m6A-positive transcripts over the total amount of input transcripts. The data represent the mean ± SD from n = 4 biological replicates. For (B) and (C), statistical significance was determined by unpaired two-tailed t-test and the P-values are indicated.