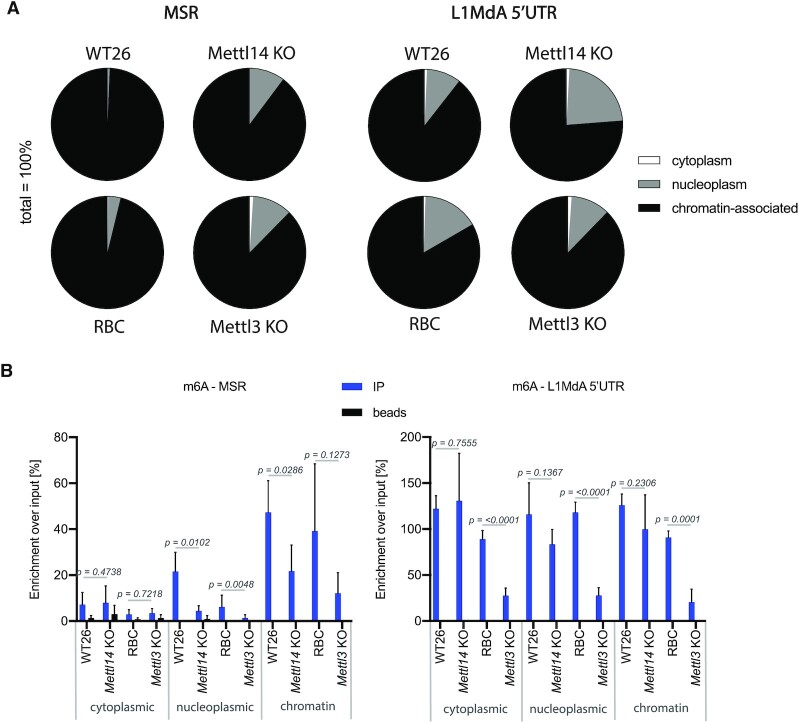
Figure 5.
Impaired chromatin association of MSR transcripts in Mettl14 and Mettl3 mutant ES cells. (A) Pie charts showing the distribution of MSR and LINE L1MdA 5′UTR transcripts in cytoplasmic (white), nucleoplasmic (grey) and chromatin (black) fractions of WT26, Mettl14 KO, RBC and Mettl3 KO ES cells. RNA was isolated from each of the subcellular fractions and MSR and LINE L1MdA 5′UTR transcripts were quantified by RT-qPCR. The relative abundance of transcripts in each subcellular fraction is displayed as the percentage of the sum of transcripts in all three subcellular fractions (100%). The data represent the mean from n = 3 biological replicates. (B) Histogram showing the m6A MeRIP enrichment for m6A-positive MSR (left) and LINE L1MdA 5′UTR (right) transcripts in RNA from cytoplasmic, nucleoplasmic and chromatin fractions of WT26, Mettl14 KO, RBC and Mettl3 KO ES cells. The data represent the mean ± SD from n = 4 biological replicates and are normalized to the input of each fraction. Statistical significance was determined by unpaired two-tailed t-test and the P-values are indicated.