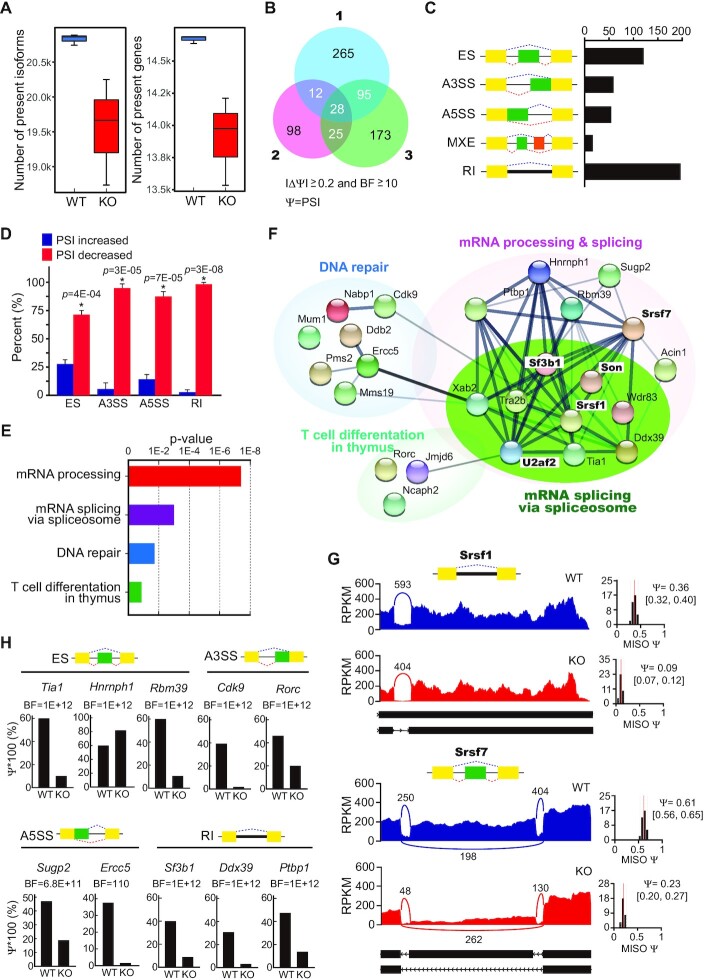
Figure 3.
Global alternative RNA splicing changes in NSrp70-cKO DP thymocytes. (A) The number of isoforms and genes present in the given samples. We checked the number of isoforms and genes present in given wild-type and KO samples and found significant reductions of the numbers of isoforms and genes in DP thymocytes (6.13% and 5.5% reductions, respectively; Wilcoxon rank sum tests, one-sided, P-values ≤ 0.05). (B) Venn diagram of the number of significantly alternative spliced-genes from three mRNA-seq datasets of Nsrp1f/f (WT) and Nsrp1f/fCD4Cre (KO) DP thymocytes. PSI, Percent spliced in; BF, Bayes’ factor. (C) Quantification of the different alternative splicing events affected by NSrp70. (D) The relative percent of each alternative splicing event affected by NSrp70. The small horizontal lines indicate the mean ± standard deviation *, meaningful P-value. (E, F) Gene ontology analysis (E) and functional association network (STRING) (F) of alternative spliced-targets affected by NSrp70-cKO. Fisher exact P-values were plotted for each enriched functional category. (G) Sashimi plot of alternative exons affected by NSrp70. Genes were chosen to represent both an increase and a decrease of PSI, and the numbers of exon junction reads are indicated. (H) PSI and BF values of different types of NSrp70-regulated alternative splicing events based on MISO analysis.