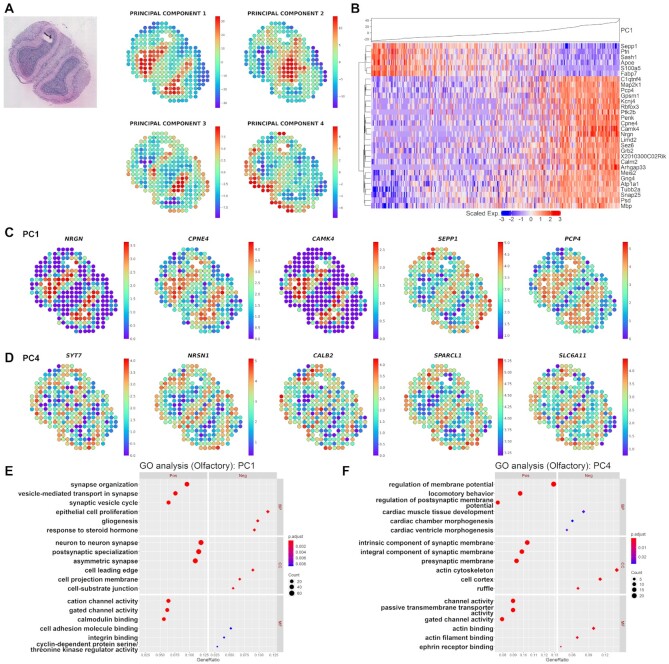
Figure 2.
Investigation of morphological marker genes and functions in olfactory bulb data. (A) Spatial mapping of the PC1, PC2, PC3 and PC4 image latents. The PC values of each spot are visualized using colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean value, respectively. (B) Heatmap for the top 30 highly associated genes for log2RC in the PC1 image latent space from olfactory bulb tissue. Hierarchical clustering was performed for the top 30 genes, and the PC1 value in each of the spots is shown at the top. Spatial expression of the top 5 genes representing the greatest contrast in the (C) PC1 and (D) PC4 image latent space from olfactory bulb tissue. The top genes are presented in descending order of |log2RC| (FDR < 0.05). The normalized gene expression level of each spot is visualized with colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean expression, respectively. Gene ontology (GO) analysis for (E) PC1 and (F) PC4 SPADE genes showing positive or negative association with PC image latent in olfactory bulb data. The top 3 positive or negative GO terms for each subcategory, biological process (BP), cellular component (CC) and molecular function (MF), are exhibited in the left and right panel, respectively. The number of overlapping genes is expressed as the size of the dot, and the Benjamini-Hochberg adjusted P-value is exhibited with a colormap.