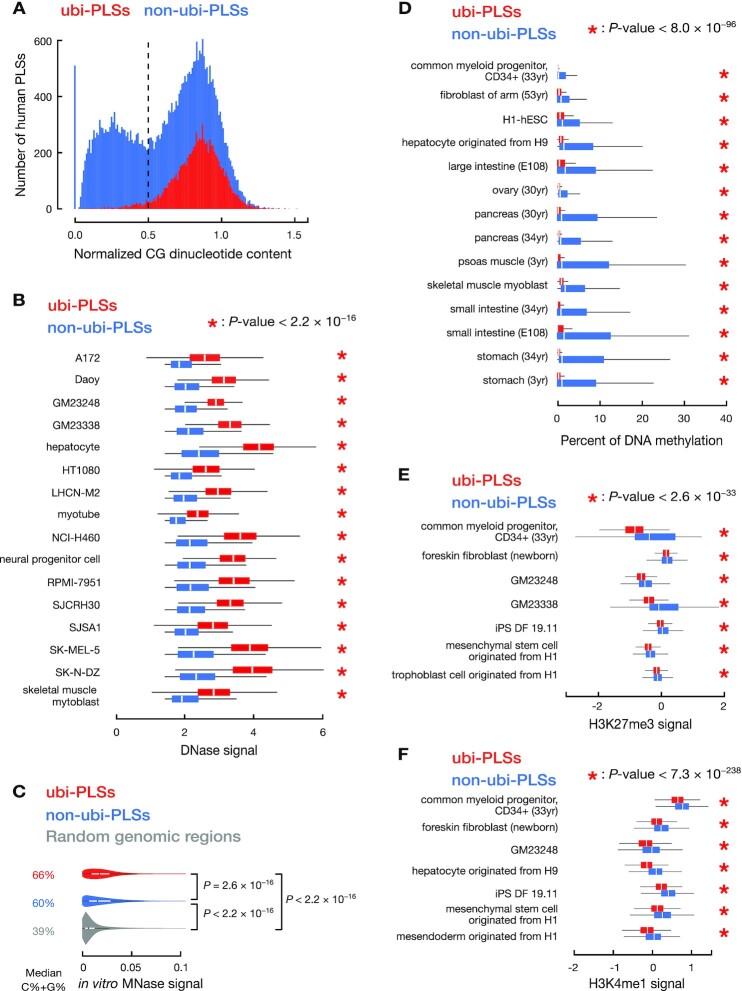
Figure 2.
ubi-PLSs are enriched in CG dinucleotides and have open chromatin in cells but closed chromatin in vitro. (A) ubi-PLSs have higher CG dinucleotide content than non-ubi-PLSs. Histograms show that a vast majority of ubi-PLSs (red) have high CG dinucleotide content (> 0.5), while non-ubi-PLSs (blue) show a bimodal distribution of CG content. (B) ubi-PLSs (red) have significantly higher DNase signals than non-ubi-PLSs (blue) defined in the same biosample. Data for 16 biosamples are shown, with each biosample represented by a pair of boxplots. All P-values were computed with Wilcoxon rank-sum tests. (C) ubi-PLSs (red) have higher in vitro nucleosome occupancy (in vitro MNase-seq signals) than non-ubi-PLSs (blue). The in vitro MNase-seq experiment was performed on in vitro reconstructed nucleosomes using purified genomic DNA and recombinant histone proteins. We randomly selected 10 000 genome regions as control (gray). All P-values were computed using Wilcoxon rank-sum tests; P-values ≤ 2.2 × 10−16 for all comparisons. (D) Similar to Figure 2B, but for DNAme levels in 14 biosamples. (E) Similar to Figure 2B, but for H3K27me3 levels in seven biosamples. (F) Similar to Figure 2B, but for H3K4me1 levels in seven biosamples.