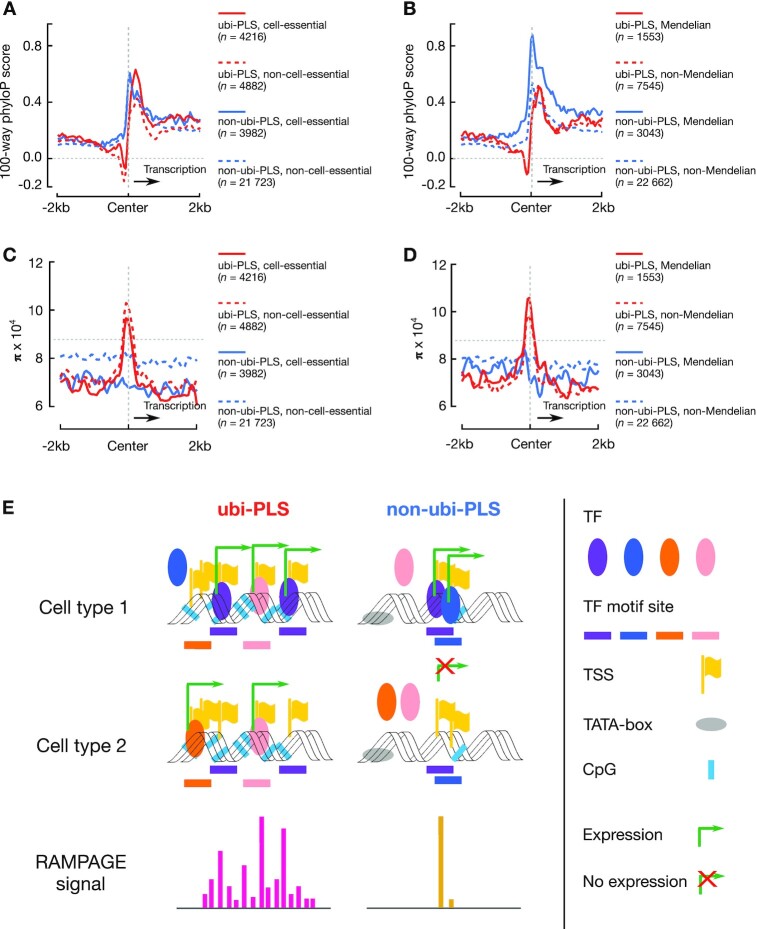
Figure 7.
Evolution conservation and human variation around ubi-PLSs and non-ubi-PLSs and a schematic model of transcriptional activation for the two groups of cCRE-PLSs. (A) Average 100-way phyloP scores in the ±2 kb genomic regions centered on human ubi-PLSs (red) and non-ubi-PLSs (blue), each subdivided into cell-essential (solid line) and non-cell-essential (dashed line) subsets. The horizontal dashed gray line represents the genome-wide background and the vertical dashed gray line indicates the center of the cCRE-PLSs in all panels A–D. (B) Average 100-way phyloP scores in the ±2 kb genomic regions centered on human ubi-PLSs (red) and non-ubi-PLSs (blue), each subdivided into Mendelian-disease (solid line) and non-Mendelian-disease (dashed line) subsets. (C) Average nucleotide diversity (π) in the ±2 kb genomic regions centered on human ubi-PLSs (red) and non-ubi-PLSs (blue), each subdivided into cell-essential (solid line) and non-cell-essential (dashed line) subsets. (D) Average nucleotide diversity (π) in the ±2 kb genomic regions centered on human ubi-PLSs (red) and non-ubi-PLSs (blue), each subdivided into Mendelian-disease (solid line) and non-Mendelian-disease (dashed line) subsets. (E) ubi-PLSs tend to have high CG content, lack a TATA-box, be bound by multiple transcription factors, and initiate transcription from dispersed genomic positions. They recruit different transcription factors to initiate transcription in different cell types, maintaining high and ubiquitous expression. In contrast, non-ubi-PLSs are more likely to have lower CG content, contain a TATA-box, be bound by fewer transcription factors, and initiate transcription from a specific genomic position. They are expressed in a few cell types and controlled by a select group of transcription factors. TF: transcription factor; TSS: transcription start site.