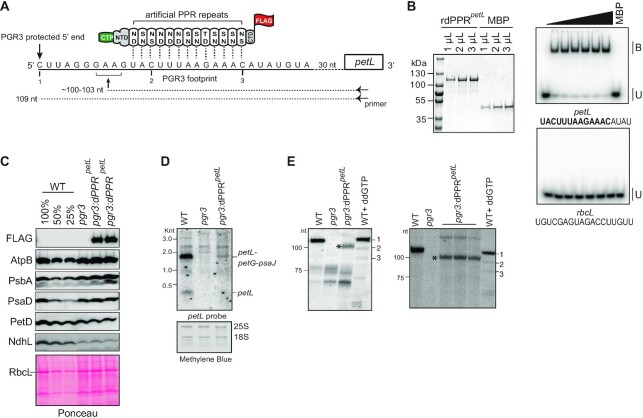
Figure 5.
Partial complementation of PGR3′s petL RNA stabilization function by dPPRpetL. (A) Schematic of dPPRpetL design. The protein consists of 13 consensus PPR motifs programmed to bind the indicated sequence in the petL 5′UTR. The amino acids at positions 5 (top) and 35 (bottom) in each dPPR motif are indicated. The dPPR tract was flanked by the N- and C-terminal regions (NTD and CTD) of maize PPR10, including PPR10’s chloroplast targeting peptide (CTP). A 3xFLAG tag was fused to the C-terminus. The 5′ end that is stabilized by PGR3 in vivo is marked with a vertical arrow, and the PGR3 footprint is underlined with a solid line. The horizontal arrow denotes the primer used for primer extension and the asterisk marks the 5′-end that is stabilized by dPPRpetL. The nucleotides marked 1, 2, and 3 are primer extension stops resulting from the use of the chain terminator ddG, which serve as size markers on the primer extension gels in panel E. (B) EMSA showing preferential binding of recombinant dPPRpetL (rdPPRpetL) to petL 5′ end. (Left) An aliquot of the purified rdPPRpetL and MBP used for binding assays was analyzed by SDS-PAGE and staining with Coomassie Blue. rdPPRpetL has a predicted size of ∼118 kDa. (Right) rdPPRpetL was used in EMSAs with radiolabeled RNA oligonucleotides (petL or rbcL) whose sequences are shown below. The sequence of the designated dPPRpetL binding site is highlighted in bold. Protein concentrations were 0, 5.6, 11.25, 22.5, 45, 90 and 118 nM for rdPPRpetLand 200 nM for the maltose binding protein (MBP). Bound (B) and unbound (U) RNAs are indicated. (C) Immunoblot analysis of pgr3 mutants expressing dPPRpetL. Plants used for this analysis were grown at low light intensity (60 μE). Results from plants grown at 120 μE are shown in Supplementary Figure S5B. Protein analyzed in each lane comes from pooled plants that were confirmed to have the indicated genotype by PCR. Replicate blots were probed with antibodies to the indicated proteins. The FLAG antibody detects dPPRpetL. The Ponceau S-stained blot is shown below to illustrate equal sample loading and the abundance of the large subunit of Rubisco (RbcL). (D) RNA gel blot hybridization demonstrating partial rescue of petL transcripts in pgr3 mutants expressing dPPRpetL. 2 μg of leaf RNA was loaded in each lane. The methylene blue stained blot is shown below to demonstrate equal sample loading. 25S and 18S are cytosolic rRNAs. (E) Primer extension assays demonstrating stabilization of the expected novel petL 5′ terminus by dPPRpetL. The two panels show analysis of four independent pools of pgr3 mutants expressing dPPRpetL. The positions of size markers are shown to the left. The last lane in each gel (WT + ddGTP) shows a primer extension reaction that used wild-type RNA and a trace amount of ddGTP to induce termination at C residues in the template (see panel A). The gel on the left used 4 μg of RNA from the WT (Ws-0) and pgr3 samples, and 14 μg from pgr3:dPPRpetL. The gel on the right used 2.5 μg of RNA from the WT (Ws-0) and pgr3 samples, and 7.5 μg from pgr3:dPPRpetL.