Fig 7. Extended mathematical model of SCN rhythm production.
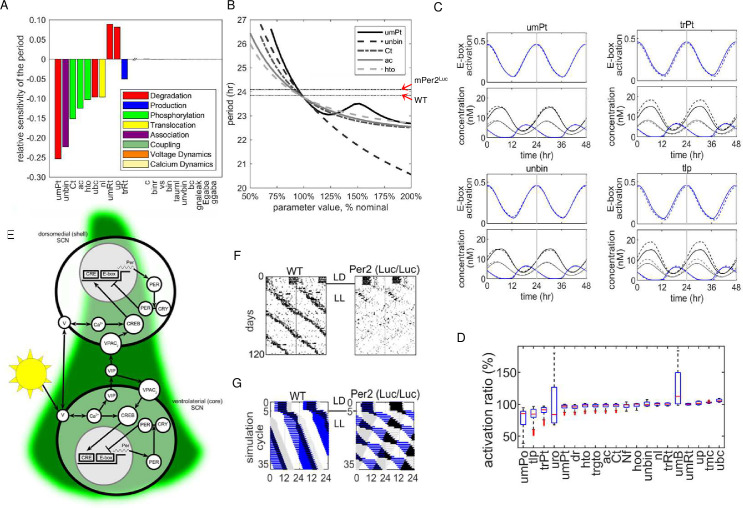
A. Parameter sensitivity analysis for single pacemaker cell period. Histograms represent the percent change for each parameter required to alter period in DD from WT to mPer2Luc/Luc. B. Sensitivity of period to the top 5 parameters. The model’s nominal base period is set at 23.8h for WT and 24.3 for mPer2Luc/Luc as indicated by the two horizontal lines. C. Circadian patterns of dependent variables represented by changes in E-box occupation/activation and total concentrations of PER1, PER2, BMAL/CLOCK as a result of changing four parameters (umPt, unbin, trPt, uro) to obtain the mutant period. Solid lines = WT model. Dashed lines = mPerLuc model. Time courses are normalized so that maximum E-box activation occurs at time = 0. Additional simulations are contained in S2A–S2H Fig. D. Comparison of E-box activation for simulations of WT and mPER2 Luc rhythms. The box and whisker plots show the distribution over one cycle: red lines are medians; blue boxes are first-to-third quartiles; black lines are extrema; and red dots are outliers. 100% = no difference between genotypes. E. Functional anatomy of the SCN pacemaker. The model is based on a primary transcription-translation feedback loop in all cells. The dynamics of the membrane potential V is coupled to the molecular clock through cytosolic calcium. The single cell model of the molecular clock is used for both the dorsal and ventral SCN regions. The regional difference is produced by the light input, and inter-regional communication is via VIP driving PER transcription through VPAC2 and the cAMP response element (CRE). VIP is produced and released from cells in the ventral SCN onto both shell and core cells. F. Representative actograms from WT and Per2Luc mice housed in LL. G. Simulations using the two oscillator SCN model in LL with baseline parameters (left) and adjustment of Per2 mRNA stability (umPt) to represent the presence of Per2Luc. Parameter symbols: umPt: degradation rate of Per2 RNA; unbin: normalized unbinding rate for BMAL1-CLOCK/NPAS2 to Per1/2/Cry1 E-box; ac: binding rate for PER1/2 to CK1ε/δ; Ct: total Ck1 concentration; hto: CK1ε/δ phosphorylation rate for PER2; ubc: degradation rate for BMAL1-CLOCK/NPAS2; nl: nuclear localization rate for proteins bound to PER; trPt: transcription rate for Per2; mRt: degradation rate for Cry2; up: degradation rate for CK1 phosphorylated PER; vs: production rate of cAMP when VPAC2R bound by VIP; dg: unbinding rate for PER2/REV-ERBs to GSK-3β; cvbin: binding rate of CRY1/2 to VPAC2R; uro: degradation rate constant for CRY1.