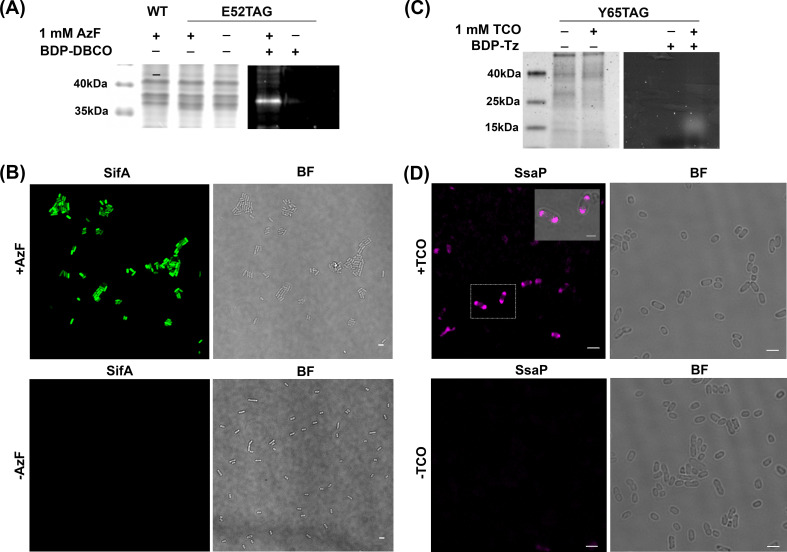
Figure 2. AzF is site-specifically incorporated into SifA-E52TAG in Salmonella.
(A) Left: Coomassie stained SDS-PAGE; right: SDS-PAGE fluorescence imaging confirms selective labeling of SifA-E52AzF with BDP-DBCO in the cell lysate. (B) Expression of SifA-E52AzF in Salmonella analyzed by fluorescence microscopy in the presence (top) or absence (bottom) of 1mM AzF. Salmonella cells expressing SifA-E52AzF in the presence or absence of AzF were incubated with BDP-DBCO, and imaged for BDP fluorescence (green). SifA fluorescence (green) is only observed in the presence of AzF. (C) Site-specific incorporation of TCO*Lys into SsaP-Y65TCO in Salmonella left: Coomassie stained SDS-PAGE; right: SDS-PAGE fluorescence imaging confirms selective labeling of SsaPY65TCO with BDP-Tz in the secreted fraction collected after TCA precipitation. (D) Expression of SsaP-Y65TCO in Salmonella analyzed by fluorescence microscopy in the presence (top) or absence (bottom) of 1mM TCO*Lys. Salmonella cells expressing SsaP-Y65TCO in the presence or absence of TCO*Lys were incubated with SiR-Tz, fixed at 8 hr post acid induction and imaged for SiR fluorescence (magenta). Polarly localized SsaP fluorescence (magenta) is only observed in the presence of TCO*Lys. A higher magnification merged imaged is shown in the inset. See Figure 2—figure supplement 5 for statistical analysis for the fraction of polarly localized SsaP. Images were acquired using confocal microscopy. The data are representative of at least three independent experiments. BF = bright field. Scale bar, 2 µm (B,D), 1 µm (inset).
Figure 2—figure supplement 1. SsaP is required for SPI-2 secretion.
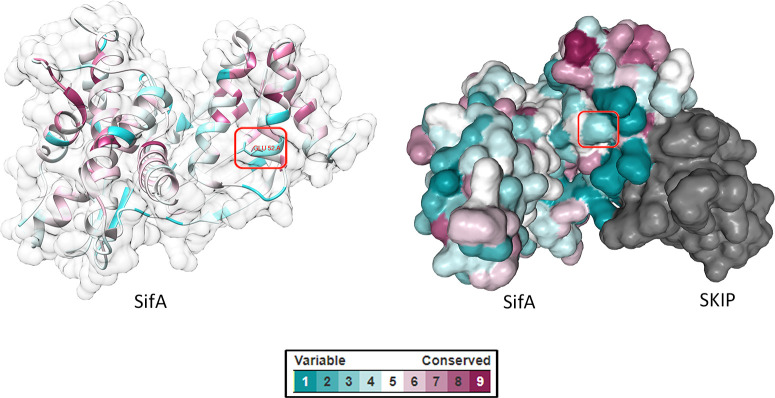
Figure 2—figure supplement 2. Selection of ncAA incorporation sites.
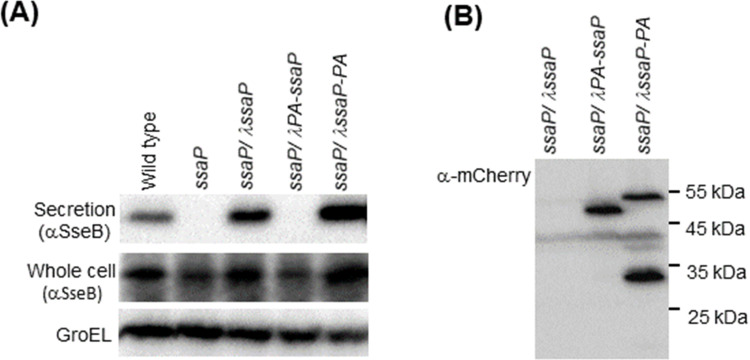
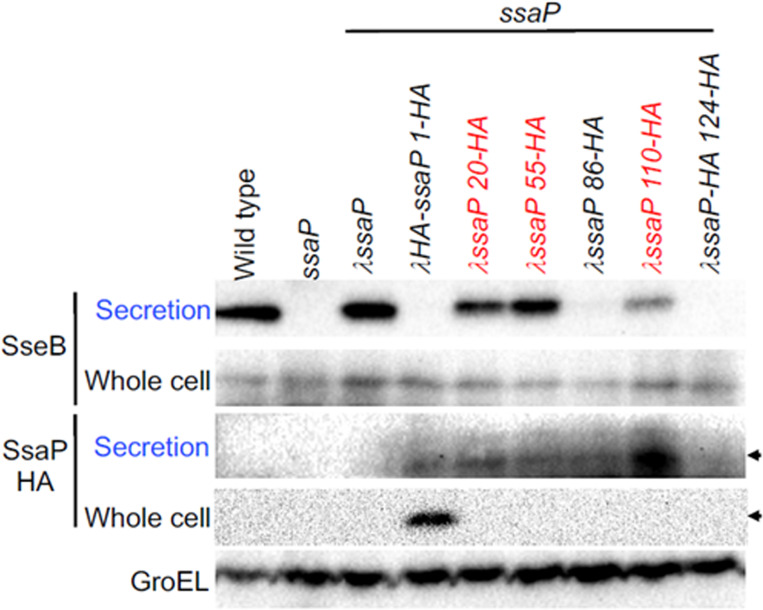
Figure 2—figure supplement 3. Western blot of SseB (translocon) and SsaP-HA secretion.
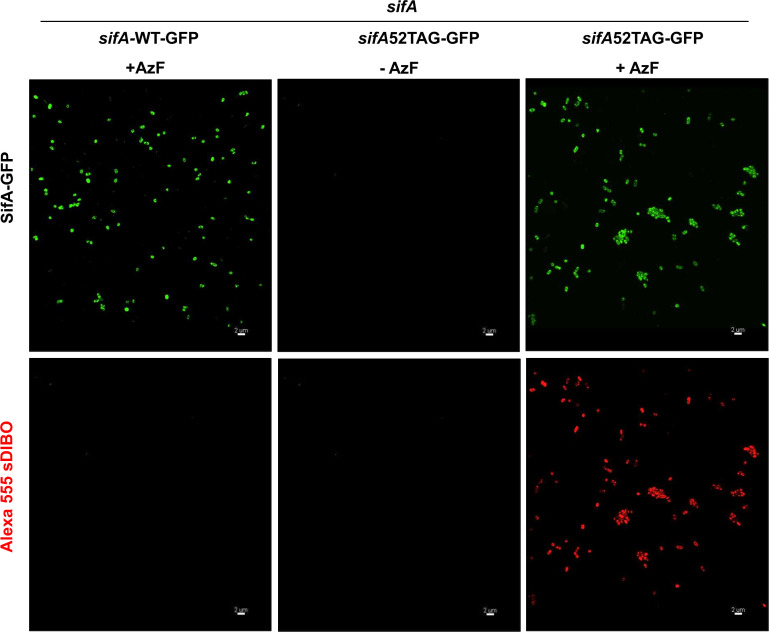
Figure 2—figure supplement 4. AzF and subsequent Alexa555 sDIBO Alkyne incorporation is highly specific.
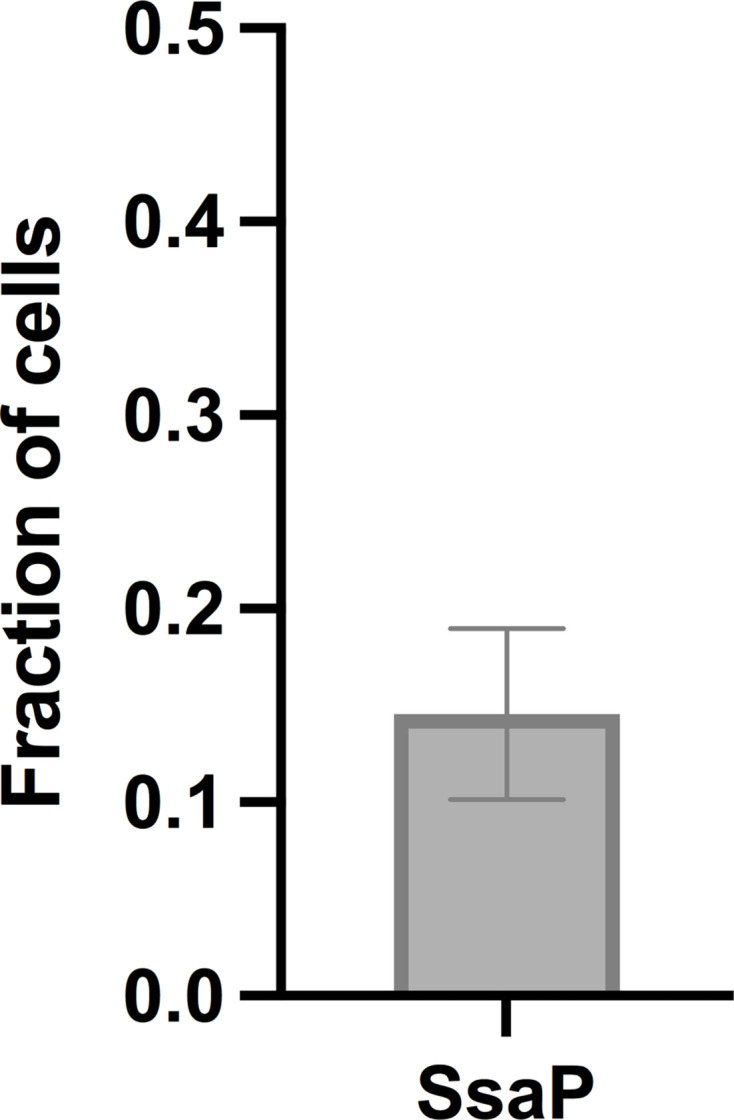
Figure 2—figure supplement 5. Average fraction of cells that display polarly localized SsaP (ntotal = 112 cells).