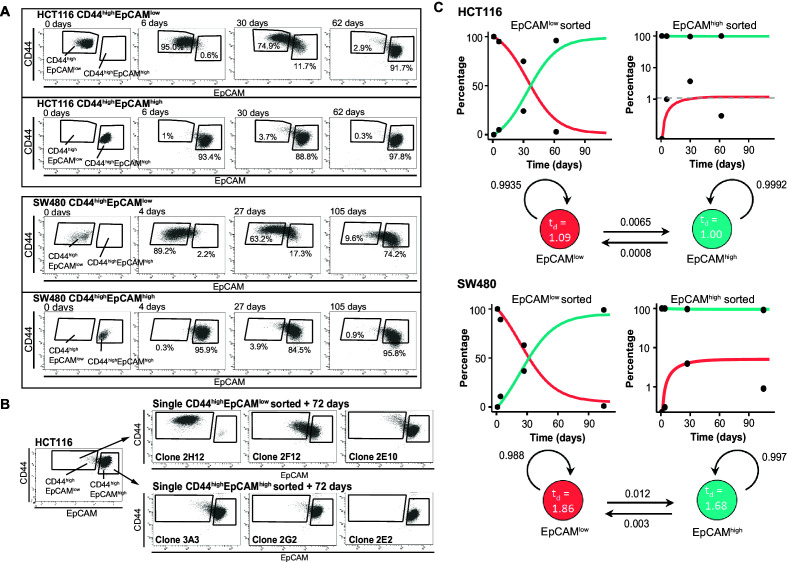
Figure 2. Phenotypic plasticity maintains EpCAMlo and EpCAMhi cells in a stochastic equilibrium.
(A) Analysis of plasticity of EpCAMhi and EpCAMlo cells from HCT116 (upper panel) and SW480 (lower panel). EpCAMhi and EpCAMlo cell fractions were sorted and plated in culture. At different time points, as indicated, cells were reanalyzed by flow cytometry for their levels of CD44 (y-axis) and EpCAM (x-axis) expression. (B) Flow cytometric analysis of single CD44hiEpCAMhi and CD44hiEpCAMlo HCT116 cells sorted by FACS and cultured for 72 days. Three representative individual single-cell clones per cell fraction are shown. (C) Dynamics of the EpCAMhi and EpCAMlo subpopulations from the HCT and SW480 cell lines as measured by FACS (% of total) over time. Under each graph, a schematic shows the estimated transition probabilities from the fitted two-state Markov model.