Extended Data Fig. 6. Allele ratios of heterozygous SNPs from CD34+ HSPC colonies after editing.
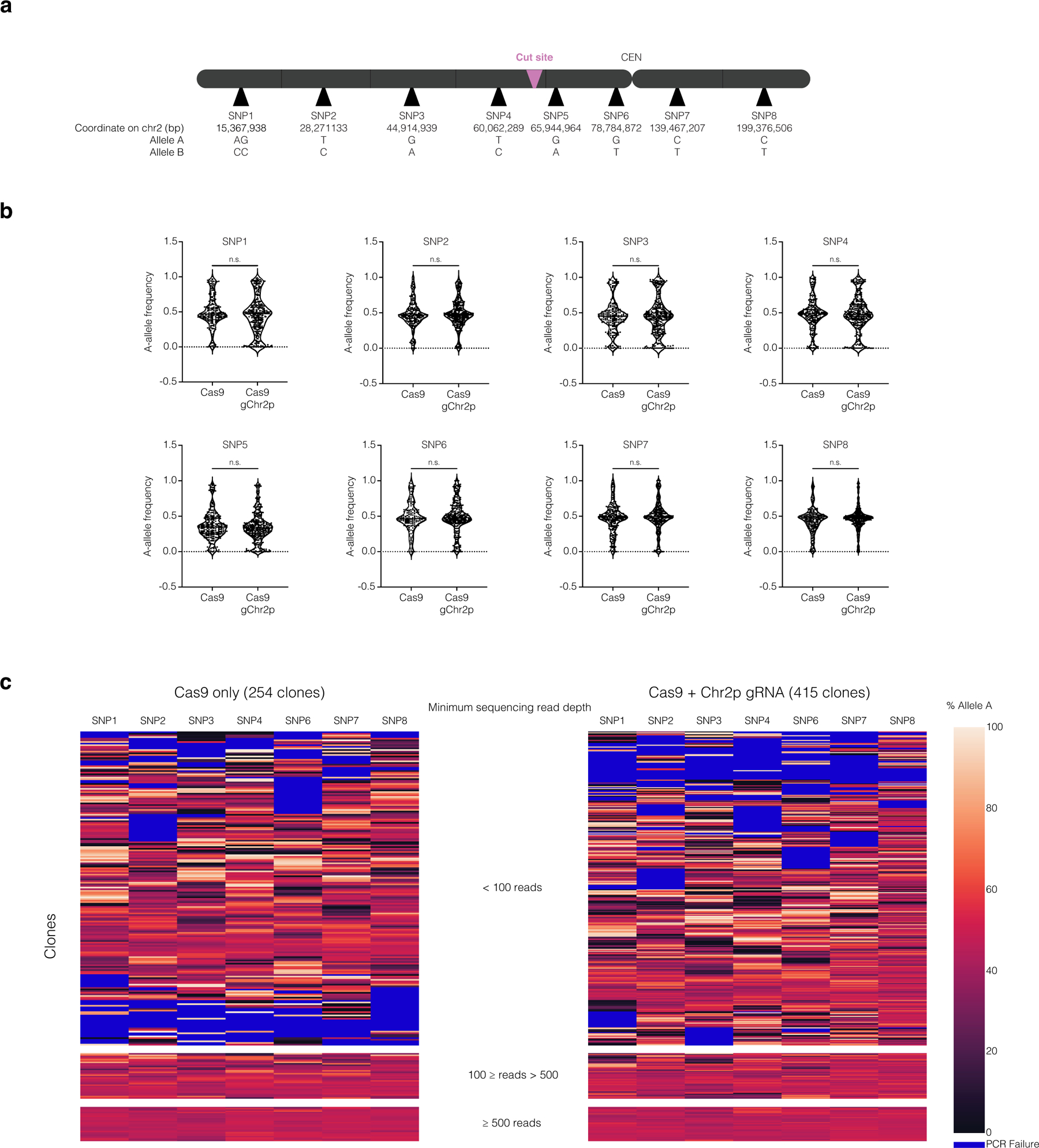
(a) Map of SNP locations, cut site, and the centromere (CEN) on chromosome 2 (not to scale).
(b) The distribution of A-allele frequencies for samples where A-allele and B-allele frequencies comprise greater than 90 % of the sequence reads. The p-values for SNPS 1–8 are p = 0.1089, 0.3140, 0.9967, 0.7792. 0.2751, 0.4659, 0.3178, and 0.2239 respectively (two-tailed Mann-Whitney U test). SNP5 exhibited a strong deviation from a 50:50 allelic ratio even in unedited controls, which may reflect a PCR amplification artifact. Because of this, SNP5 was excluded from subsequent analysis.
(c) Heatmap of allele frequency data for all samples (Cas9, left; Cas9 + Chr2p gRNA, right). The heatmap is divided into sections based on the minimum sequencing read depth. Minimum sequencing read depth was defined by the SNP with the lowest number of reads in the sample. Samples with low read depth exhibited high variability in allelic ratios, likely reflecting low input DNA from small colonies. Because we lack phasing information, any deviation from a 50:50 allele ratio for multiple adjacent SNPs suggests segmental copy number alterations.
See Supplementary Note for methods and additional discussion. For this experiment, only several hundred clones could feasibly be grown and analyzed, whereas patients will receive tens to hundreds of millions of edited cells. From the several hundred clones in our experiment, we only expect ~20 cells containing micronuclei based on micronucleation rates measured in Fig. 6. Extrapolating from these data, patients will receive millions of micronucleated cells, each one with the potential to undergo chromothripsis and grow into a clone. We note that this assay will not detect copy-number neutral chromothripsis nor chromothripsis that maintains copy number and heterozygosity at the assayed SNPs, with rearrangements located on other segments of the edited chromosome. Moreover, this approach has a limited ability to detect copy number gains or subclonal events that result from ongoing genomic instability triggered by micronucleation or bridging derived from the initial editing.
