Figure 6. Hallmark cytological features of chromothripsis after a genome editing approach for the treatment of sickle cell disease.
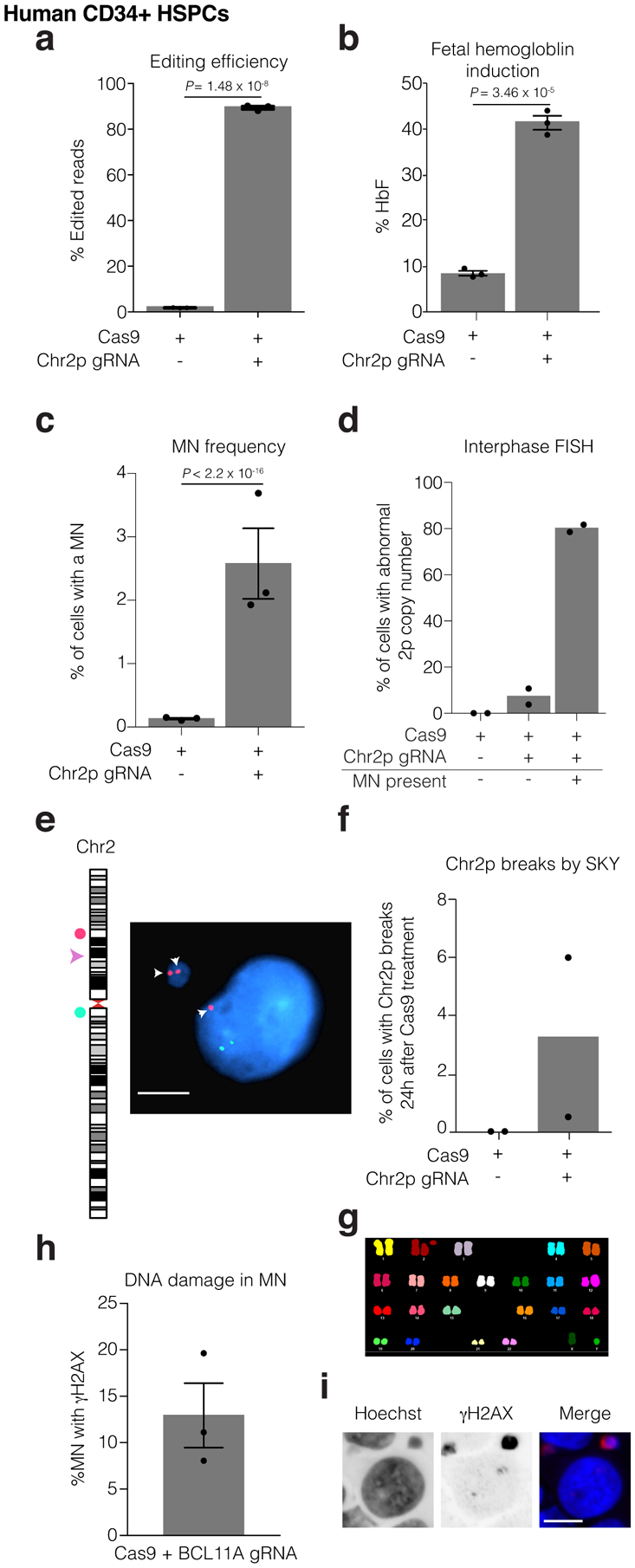
Human CD34+ HSPCs were electroporated with Cas9/gRNA RNP targeting the erythroid-specific enhancer of BCL11A. Microscopic analysis of micronucleation was performed 24 h post electroporation.
(a) Editing efficiency of BCL11A determined from amplicon sequencing. n = 3 experiments, Error bars: mean +/− SEM, two-tailed unpaired t-test).
(b) Fetal hemoglobin (HbF) levels were measured by HPLC in erythroid-differentiated CD34+ HSPCs as a functional readout of successful editing of BCL11A 10 days after RNP electroporation. n = 3 experiments, Error bars: mean +/− SEM, two-tailed unpaired t-test).
(c) Percent of cells with a micronucleus (n = 3 experiments with 7827 and 6480 cells counted, left to right). Error bars: mean +/− SEM, two-tailed Fisher’s exact test.
(d) Percent of cells with aberrant 2p copy number assayed by FISH (n = 2 experiments with 1957, 1926, 74, cells counted, left to right).
(e) Representative FISH image of data in (d). Cut site is represented by a pink arrowhead; DNA is blue; telomere proximal probe is red, and marked by arrows; centromere proximal probe is green. Shown is a micronucleated cell with 3 copies of the cut arm, two of which are in the micronucleus. Scale bar 5 μm.
(f) Chr2p breaks present 24 hours after electroporation in metaphase visualized by SKY (n = 2 experiments, 400 spreads per condition).
(g) Sample SKY image from (f).
(h) Percent of CD34+ CRISPR-MN with extensive DNA damage covering the DNA present in the micronucleus by γH2AX-labeling (n = 3 experiments, 135 micronuclei scored). Error bars: mean +/− SEM.
(i) Representative image of data in (h). Scale bar 5 μm.
