Extended Data Fig. 2. DNA damage, nuclear envelope rupture and reduced DNA replication in CRISPR-MN.
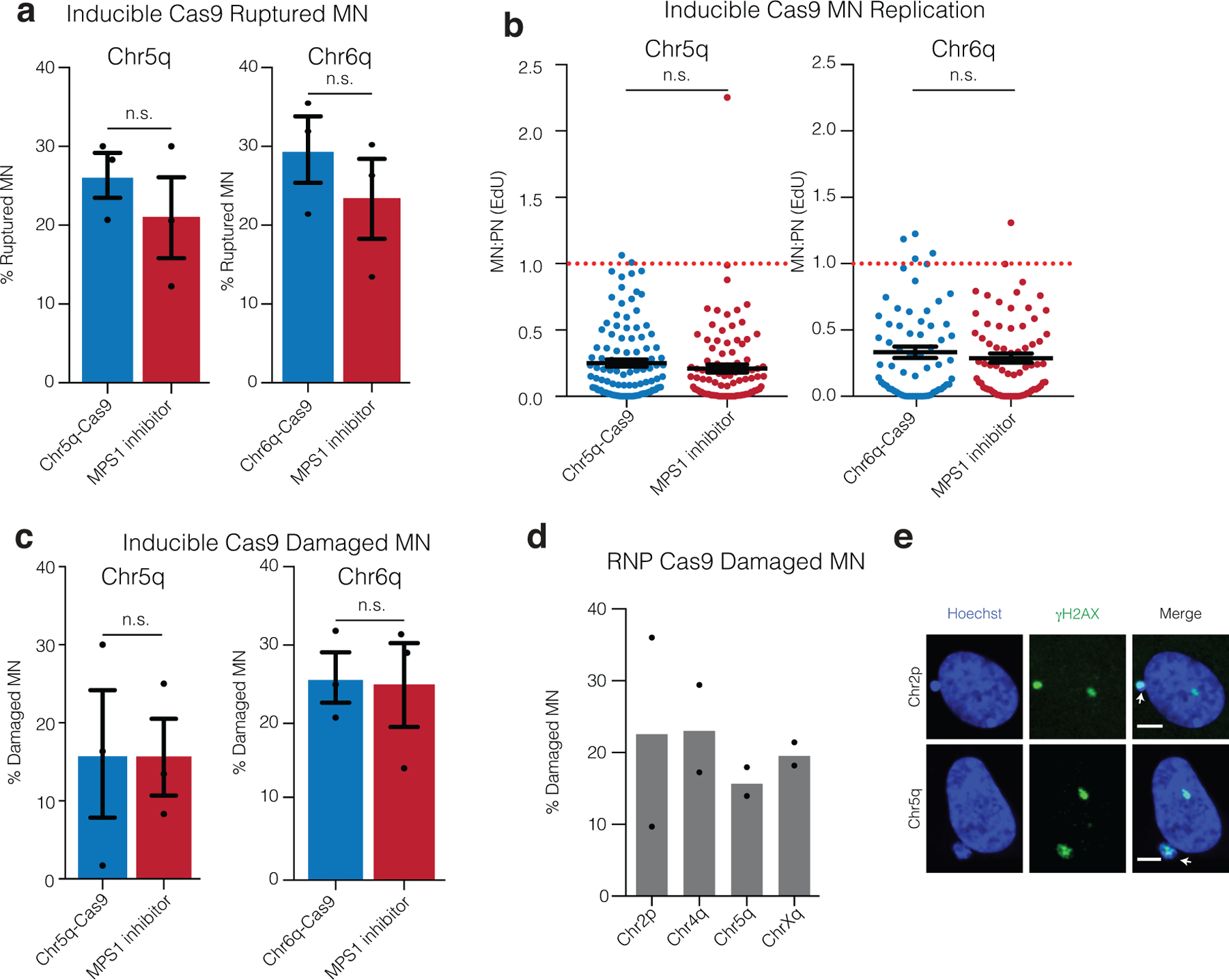
(a) Nuclear envelope rupture frequency for CRISPR-MN as compared to spindle checkpoint inhibitor-induced micronuclei. Rupture was defined as an MN:PN ratio of lamin B receptor (LBR)49 intensity > 3 (n = 3 experiments with 201 and 167 micronuclei analyzed for chr5q, p = 0.2216 and 165 and 152 micronuclei counted for chr6q, p = 0.2034). Error bars: mean +/− SEM, two-tailed Fisher’s exact test.
(b) DNA replication defect of CRISPR-MN. EdU fluorescence intensity was measured after a 5-hour pulse. Only cells that had entered S-phase were scored (>150 a.u. EdU signal in primary nucleus). Dotted red line is normal levels of DNA replication in the micronucleus relative to the primary nucleus (n = 3 experiments with 109 and 97 micronucleated cells analyzed for chr5q, p = 0.1698 and 65 and 73 micronucleated cells analyzed for chr6q, p = 0.6948). Error bars: mean +/− SEM; two-tailed Mann-Whitney U-test.
(c) CRISPR-MN acquire DNA damage. Shown is the frequency of γH2AX positive micronuclei (> 3 standard deviations above mean signal in primary nuclei) for the indicated gRNAs using the inducible Cas9 system (n = 3 experiments with 203 and 184 micronucleated cells analyzed for chr5q, p = 0.6870 and 175 and 169 cells analyzed for chr6q, p = 0.8053). Error bars: mean +/− SEM, two-tailed Fisher’s exact test.
(d) CRISPR-MN acquire DNA damage (RNP Cas9 system). Shown is the frequency of γH2AX positive micronuclei for the indicated gRNAs (n = 2 experiments with 56, 46, 82, and 50 micronucleated cells analyzed, left to right).
(e) Example images of data from panel (d) showing γH2AX labeling. White arrows: micronuclei. Scale bars, 5 μm. The γH2AX focus in the primary nucleus likely decorates the centric portion of the broken chromosome. Alternatively, or additionally, it may label a DNA break on the homolog.
